Abstract
Rosa rugosa is an excellent landscaping plant famous for its fragrance and primarily red flower. In this study, transcriptome sequencing on R. rugosa ‘Red Zizhi’ and its bud mutations R. rugosa ‘Pink Zizhi’ and R. rugosa ‘White Zizhi’ were conducted using the Illumina HiSeq™ 2000 platform. In total, 103,446 assembled contigs and 51,332 unigenes were obtained with average lengths of 359 and 735 bp, respectively. A total of 40,018 (77.96%) unigenes were functionally annotated; 20,054 (39.07%) of which were annotated in 128 Kyoto Encyclopedia of Genes and Genomes database pathways. Moreover, 316 (1.58%) unigenes were annotated to phenylalanine synthetic pathways related to coloration, 191 (0.95%) to flavonoid synthetic pathways and 15 (0.07%) to anthocyanin synthetic pathways. Analysis of differential gene expression profiles showed that ‘White Zizhi’ and ‘Pink Zizhi’ differed by 431 unigenes, ‘Red Zizhi’, and ‘Pink Zizhi’ by 1319 unigenes, and ‘Red Zizhi’ and ‘White Zizhi’ by 1324 unigenes. Five candidate genes were possibly highly related to the biosynthesis of rose flower pigments; they were selected by applying the differentially expressed gene method. Analyses were conducted to confirm the expression patterns of selected genes and changes and contents of flower pigments in petals of the three rose varieties in different flower development stages. Finally, by analyzing the relationship between gene expression and flower pigment synthesis, we discovered that the 4-coumarate:CoA ligase gene is possibly related to the formation of pink flowers, and low dihydroflavonol 4-reductase gene expression levels may be related to color formation in ‘White Zizhi’.






Similar content being viewed by others
Abbreviations
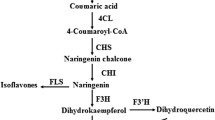
- 4CL:
-
4-Coumarate:CoA ligase gene
- CHS:
-
Chalcone synthase
- CHI:
-
Chalcone isomerase
- DHK:
-
Dihydrokaempferol
- DHQ:
-
Dihydroquercetin
- DHM:
-
Dihydromyricetin
- DFR:
-
Dihydroflavonol 4-reductase
- UA3′5′GT:
-
UDP-glucose:anthocyanin 3′,5′-O-glucosyltransferase
- DGE:
-
Digital gene expression
- DEGs:
-
Differentially expressed genes
- ESI:
-
Electrospray ionization
- HPLC:
-
High-performance liquid chromatography
- MS:
-
Mass spectrometry
- CDS:
-
Coding DNA sequence
- NR:
-
Non-redundant protein database
- Swiss-Prot:
-
Swiss-Prot protein database
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes database
- COG:
-
Cluster of orthologous groups of proteins database
- NT:
-
Non-redundant nucleotide database
- GO:
-
Gene ontology
References
Alagna F, D’Agostino N, Torchia L, Servili M, Rao R, Pietrella M, Giuliano G, Chiusano ML, Baldoni L, Perrotta G (2009) Comparative 454 pyrosequencing of transcripts from two olive genotypes during fruit development. BMC Genom 10(34):399
Allina SM, Prihadash A, Theilmann DA, Ellis BE, Douglas CJ (1998) 4-coumarate: coenzyme a ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes. Plant Physiol 116(2):743–754
Barakat A, Diloreto DS, Zhang Y, Smith C, Baier K, Powell WA, Wheeler N, Sederoff R, Carlson JE (2009) Comparison of the transcriptomes of american chestnut (Castanea dentata) and chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biol 9:51
Chen S, Luo H, Li Y, Sun Y, Wu Q, Niu Y, Song J, Lv A, Zhu Y, Sun C, Steinmetz A, Qian Z (2011) 454 EST analysis detects genes putatively involved in ginsenoside biosynthesis in Panax ginseng. Plant Cell Rep 30(9):1593–1601
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2go: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676
Feng LG, Chen C, Li TL, Wang M, Tao J, Zhao DQ, Sheng LX (2014) Flowery odor formation revealed by differential expression of monoterpene biosynthetic genes and monoterpene accumulation in rose (Rosa rugosa Thunb.). Plant Physiol Bioch 75:80–88
Franssen SU, Shrestha RP, Bräutigam A, Bornberg-Bauer E, Weber AP (2011) Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genom 12(5):227
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652
Graham IA, Besser K, Blumer S, Branigan CA, Czechowski T, Elias L, Guterman I, Harvey D, Isaac PG, Khan AM, Larson TR, Li Y, Pawson T, Penfield T, Rae AM, Rathbone DA, Reid S, Ross J, Swallwood MF, Segura V, Townsend T, Vyas D, Winzer T, Bowles D (2010) The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science 327(5963):328–331
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57(1):761–780
Hao DC, Ge GB, Xiao PG, Zhang YY, Yang Ling (2011) The first insight into the tissue specific taxus transcriptome via illumina second generation sequencing. PLoS One 6(6):e21220
Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Bio 1999:138–148
Jewett MC, Oliveira AP, Patil KR, Nielsen J (2005) The role of high-throughput transcriptome analysis in metabolic engineering. Biotechnol Bioproc E 10(5):385–399
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10(5):236–242
Kogawa K, Kato N, Kazuma K, Noda N, Suzuki M (2007) Purification and characterization of UDP-glucose: anthocyanin 3′,5′-O-glucosyltransferase from Clitoria ternatea. Planta 226(6):1501–1509
Krol ARVD, Mur LA, Lange PD, Mol JNM, Stuitje AR (1990) Inhibition of flower pigmentation by antisense CHS genes: promoter and minimal sequence requirements for the antisense effect. Plant Mol Biol 14(4):457–466
Li CQ, Wang Y, Huang XM, Li J, Wang HC, Li JG (2013) De novo assembly and characterization of fruit transcriptome in Litchi chinensis, sonn and analysis of differentially regulated genes in fruit in response to shading. BMC Genom 14(1):21–43
Liang Y, Chen SY (2011) Application of next generation sequencing techniques in plant transcriptome. Hereditas 33(12):1317–1326
Logacheva MD, Kasianov AS, Vinogradov DV, Samigullin TH, Gelfand MS, Makeev VJ, Penin AA (2011) De novo sequencing and characterization of floral transcriptome in two species of buckwheat (Fagopyrum). BMC Genom 12:30
Luo P, Ning GG, Wang Z, Shen YX, Jin HA, Li PH, Huang SS, Zhao J, Bao MZ (2016) Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front. Plant Sci 6:1257
Mikanagi Y, Saito N, Yokoi M, Tatsuzawa F (2000) Anthocyanins in flowers of genus Rosa, sections Cinnamomeae(= Rosa), Chinenses, Gallicanae, and some modern garden roses. Biochem Syst Ecol 28(9):887–902
Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628
Nakamura N, Katsumoto Y, Brugliera F, Demelis L, Nakajima D, Suzuki H, Tanaka Y (2015) Flower color modification in Rosa hybrida by expressing the S-adenosylmethionine: anthocyanin 3′,5′- O-methyltransferase gene from Torenia hybrida. Plant Biotechnol 32:109–117
Ness RW, Siol M, Barrett SC (2011) De novo sequence assembly and characterization of the floral transcriptome in cross- and self-fertilizing plants. BMC Genom 12(12):298
Nguyen TT, Guillarme D, Rudaz S, Veuthey JL (2006) Chromatographic behaviour and comparison of column packed with sub-2 μm stationary phases in liquid chromatography. J Chromatogr A 1128(1–2):105–113
Nishihara M, Nakatsuka T (2011) Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol Lett 33(3):433–441
Nováková L, Matysová L, Solich P (2006) Advantages of application of UPLC in pharmaceutical analysis. Talanta 68(3):908–918
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3(6):1101–1108
Tanaka Y, Ohmiya A (2008) Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr Opin Biotech 19(2):190–197
Tanaka Y, Tsuda S, Kusumi T (1998) Metabolic engineering to modify flower color. Plant Cell Physiol 39(11):1119–1126
Tatsuzawa F, Saito N, Shinoda K, Shigihara A, Honda T (2006) Acylated cyanidin 3-sambubioside-5-glucosides in three garden plants of the cruciferae. Phytochemistry 67(12):1287–1295
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by rna-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Wang ZY, Fang BP, Chen JY, Zhang XJ, Luo ZX, Huang LF, Chen XL, Li YJ (2010) De novo assembly and characterization of root transcriptome using illumina paired-end sequencing and development of cSSR markers in sweetpotato (Ipomoea batatas). BMC Genom 11(53):726
Wang HB, Zou ZR, Wang SS, Gong M (2013) Global analysis of transcriptome responses and gene expression profiles to cold stress of Jatropha curcas L. PLoS One 8(12):e82817
Wu Q, Sun C, Luo H, Li Y, Niu Y, Sun Y, Lu A, Chen S (2010a) Transcriptome analysis of Taxus cuspidata needles based on 454 pyrosequencing. Planta Med 77(4):394–400
Wu Q, Song JY, Sun YQ, Suo FM, Li C, Luo HM, Liu Y, Li Y, Zhang XW, Yao H, Li XW, Hu SN, Sun C (2010b) Transcript profiles of Panax quinquefolius from flower, leaf and root bring new insights into genes related to ginsenosides biosynthesis and transcriptional regulation. Physiol Plant 138(2):134–149
Ye J, Fang L, Zheng HK, Zhang Y, Chen J, Zhang ZJ, Wang J, Li ST, Li RQ, Bolund L, Wang J (2006) Wego: a web tool for plotting go annotations. Nucleic Acids Res 34(suppl 2):293-197
Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, Malerba G, Bellin D, Pezzotti M, Delledonne M (2010) Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol 152(4):1787–1795
Acknowledgements
This research was funded by the National Natural Science Foundation of China (Grant Nos. 31370696 and 31772340), Key Research and Development Program of Jiangsu Province (Modern Agriculture, Grant No. BE2015339), National Natural Science Foundation of Jiangsu Province (Grant No. BK20171284), Postgraduate Training and Innovation Project of Jiangsu Province (Grant No. KYCX17_1887), and Innovation Fund for Science and Technology of Yangzhou University (Grant No. 2016CXJ063).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by A Chandra.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sheng, L., Xia, W., Zang, S. et al. Transcriptome-sequencing analyses reveal putative genes related to flower color variation in Chinese Rosa rugosa. Acta Physiol Plant 40, 62 (2018). https://doi.org/10.1007/s11738-018-2635-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-018-2635-6