Abstract
Sirtuins are proteins belonging to the group of NADH-dependent deacetylase and mono-ADP-ribosyltransferase enzymes. Sirtuins have been discovered for the first time in yeasts, subsequent studies have shown their presence in bacteria, plants and animals. These enzymes are frequently called longevity enzymes due to the fact that they are part of genetic apparatus involved in aging control. In animals, sirtuins are key regulators of cell defense in response to stress caused by many metabolic processes; they are also involved in the regulation of cell division, metabolism, gene silencing and genetic material repair as well as apoptosis. Thus far, only several well-known research teams have been studying plant proteins resembling animal sirtuins. Considering the fact how essential functions sirtuins play in other organisms, it is extremely interesting to understand their role in plants, especially that the knowledge about them is still limited. It is believed that the function of sirtuins in Arabidopsis thaliana is associated with mitochondrial energy metabolism. Possibly they may also control the synthesis of auxins or proteins involved in their transport, or they may be responsible for regulating cellular response to auxin action. In rice, sirtuins are necessary for the protection against genomic instability and cell damage that guarantee their growth. They also take part in a defensive response against Pseudomonas syringae. They may also be involved in the ripening of fruits. Moreover, their functions are associated with photosynthetic activity and aging of leaves.
Similar content being viewed by others
Introduction
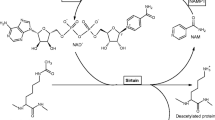
Sirtuins (SIR2 proteins—silent information regulator) belong to the third family of NADH-dependent deacetylases from the HDAC group (histone deacetylases). These enzymes catalyze the deacetylation reaction by recognizing lysine residues in ε-N-acetylated protein sequences, which results in the formation of deacetylated lysine. This lysine is located within the N-terminal domains of histone H3 and H4, and its deacetylation increases folding of these proteins, which assume more compact structure. The consequence is the formation of genetically inactive heterochromatin. In addition, sirtuins catalyze specific two-stage type of deacetylation of non-histone proteins, in the initial step of which, the hydrolysis of the glycosidic bond takes place, which links nicotinamide with ADP-ribose residue present in the NAD+ molecule. In the next step, the acyl group is transferred from the bound substrate protein to the ADP-ribose residue with a parallel formation of the 2′-and 3′-O-acetyl-ADP-ribose and release of the nicotinamide metabolite. Additionally, sirtuin belongs to the class of mono-ADP-ribosyltransferases that transfer ADP-ribose residues on protein substrates (Siedlecka and Bogusławski 2005; Shoba et al. 2009; Wang et al. 2010).
Sirtuins were discovered for the first time in yeasts and described as regulators of transcriptional gene silencing involved in conjugation as well as telomere and ribosomal DNA activation (rDNA) (Houtkooper et al. 2012). The role of SIR2 in yeasts is associated with the regulation of aging rate, and thus with extending the life span. This is related to the fact that sirtuins prevent recombination of genetic material, leading to a reduction of stem cell divisions, which is a factor limiting the life span of these organisms. Subsequent studies have shown that sirtuins are also present in bacteria, plants and animals (Siedlecka and Bogusławski 2005; Busconi et al. 2009) and play various functions. In eukaryotes, they affect cellular metabolism by engaging in transcription repression, recombination, cell cycle, and microtubule organization (Busconi et al. 2009; Mulligan et al. 2011). They are also involved in DNA repair by non-homologous end joining (NHEJ).
Classification, structure and function of sirtuins
Sirtuins have been described in various organisms, i.e., bacteria (e.g., Sulfolobus solfataricus L. major), yeasts (Saccharomyces cerevisiae), nematodes (e.g., Caenorhabditis elegans), fruit flies (Drosophila melanogaster), humans (Homo sapiens), and among plants in rice (Oryza sativa), thale cress (Arabidopsis thaliana), vine (Vitis vinifera L.) and tomato (Solanum lycopersicum). The name SIR2 is used in case of sirtuins which are enzymes present in S. cerevisiae, Drosophila and Ascaris, while mammalian sirtuin homologs are named SIRT1, SIRT2, SIRT3, etc., in plants these proteins are abbreviated SRT.
All the above-listed sirtuins are interconnected by a significant amino acid sequence similarity, which is expressed in the presence of a similar sequence motif that is flanked by N- and C-terminal extensions. This motif is conservative, consisted of 275 amino acids, which form the core of the catalytic NAD+-binding domain (referred to as PF02146), and mutations within it can lead to inhibition of the enzymatic activity of sirtuins (Grozinger et al. 2001). In turn, extensions are variable in length and sequence, and they are the target of post-translational protein modifications, during which sirtuin provides them with different functions and their subcellular localization is determined.
The larger sirtuin domain has a conformation called the Rossmann fold, a structure characteristic of proteins binding NAD+. The smaller sirtuin domain is formed by connecting two modules located in the Rossmann fold domain: zinc finger motif and a variable α-helical region (Moniot et al. 2012).
SIR2 eukaryotic proteins as well as the genes encoding them have been divided into four main classes (I–IV) based on the phylogenetic analysis of the conserved catalytic domain. The fifth class has been also distinguished, specific only for prokaryotes; however, the majority of prokaryotic sirtuins are classified as class I and III proteins (Pandey et al. 2002). The yeast SIR2 belongs to class I and is involved in chromatin silencing, DNA damage repair and chromosome condensation during meiosis (Huang et al. 2007). In D. melanogaster, it was demonstrated that the increased expression of genes encoding sirtuins may affect the length of their life cycle (Whitaker et al. 2013). In C. elegans, four genes have been identified, encoding proteins related to yeast sirtuins. The nucleotide sequence of the SIR2.1 gene present in nematodes showed 31 % similarity to the yeast SIR2 sequence, encoding conserved catalytic domain of sirtuin protein family. Other nematode genes (SIR2.2–SIR2.4) showed 10–20 % similarity to the yeast counterparts with respect to the analyzed sequence (Tissenbaum and Guarente 2001). Presumably, the function of SIR2 proteins in nematodes may be related to the level of food availability. Most probably, they are involved in the regulation of insulin-like signaling pathway. Seven members of the SIR2 protein family (SIRT1–SIRT7) have been distinguished in mammals. SIRT1, SIRT2, SIRT3 belong to class I, SIRT4 to class II, SIRT5 to III and SIRT6 and SIRT7 to class IV (Frye 2000; Houtkooper et al. 2012). They differ in cell localization, tissue localization, enzymatic activity, substrates and biological implications (Huang et al. 2007; Shoba et al. 2009; Houtkooper et al. 2012; Whitaker et al. 2013; König et al. 2014; Kupis et al. 2016) (Table 1).
So far, there are few reports concerning plant sirtuins. The research on this family of proteins in plants carried out thus far allowed to classify them into two of the four sirtuin classes previously determined in animals, namely II and IV. SRT1, a protein SIRT4-like clustered in class II, and SRT2, a protein SIRT6/SIRT7-like clustered in class IV. It was shown that both SRT1 and SRT2 have deacetylase activity. The fact that so few plant sirtuins have been discovered, while they possess multiple functions, might suggest that during the process of evolution some sirtuin genes have been lost, and their loss could have been compensated by the acquisition of new functions by the remaining members of the plant sirtuin family members. In addition, the multiplicity and diversity of such different functions played by the plant sirtuins can be explained by an example of Arabidopsis, in which different variants of the AtSRT2t gene are formed as a result of alternative splicing.
Moreover, in plants a unique family of HD2 proteins has been distinguished, which is absent in fungi or animals. These proteins have deacetylase activity, thus it cannot be ruled out that the HD2 protein took over part of the functions found in animal sirtuins, and perhaps this is the reason why in plants only two of the four classes of these proteins occurring in animals are found.
As animal proteins, plant sirtuins, in addition to the various functions, are also characterized by a different cellular localization. Huang et al. (2007) showed that SRT1 was present in mitochondria, while the study of Wang et al. (2010), Cucurachi et al. (2012) and Zhao et al. (2015) demonstrated its nuclear localization; SRT2 in all plant species studied was localized mostly in mitochondria (inner mitochondrial membrane). However, the study by Zhao et al. (2015) indicated that the homolog of Arabidopsis SRT2 in tomato was localized in both the nucleus and the cytosol. In turn, Cucurachi et al. (2012) in vine showed the presence of SRT2 in the stroma of chloroplasts and mitochondria (Table 2).
Sirtuins in animals
Sirtuins in animals are called proteins or enzymes of longevity. They are believed to be crucial in the regulation of life span, cell survival, apoptosis and metabolism in a variety of organisms. Hence, sirtuins act as factors that regulate the rate of aging, and thus they can affect the life span.
The best known sirtuins belongs to the first class, while the function of class II enzymes is the least known. The role of SIRT1 has been analyzed, i.a., during a calorie deficit, protection against old age diseases and in metabolic homeostasis (Houtkooper et al. 2012). Initially, it was discovered that SIRT1 exhibits histone deacetylase activity, but later studies showed that this activity is also directed towards other non-histone proteins, e.g., p53 or NFκB. It was shown that SIRT1 could delay aging processes by deacetylation of peroxisome proliferator-activated receptor gamma co-activator 1 alpha (PGC-1α), which is the main regulator of mitochondrial biogenesis. This protein is also involved in DNA damage repair. In neurons, it plays a role in DSBR (double-strand break repair) via NHEJ, while in proliferating cells, SIRT1 is involved in homologous recombination and base excision repair (BER) (Fang et al. 2016). Yamamori et al. (2010) proved that human AP endonucelase 1 (APE1) is a target for deacetylation by SIRT1, a protein that is directly involved in the repair of damaged DNA. Moreover, SIRT1 stimulates endonuclease activity of cellular activate protein (AP).
Further, Li et al. (2008) showed that its activity is connected with a genetic disease known as a syndrome of premature aging (Werner syndrome). In animals, SIRT1 can activate autophagy by deacetylation of light chain 3 (LC3). They can also influence the mitophagy through the regulation of mitochondrial membrane potential by maintaining the integrity of PTEN-induced putative kinase 1 (PINK1) (Fang et al. 2014).
SIRT3 is a major mitochondrial deacetylase, the activity of which has been observed in LCAD proteins (acyl-CoA dehydrogenases), involved in fatty acid β-oxidation. SIRT3 deacetylases are components I, II and III of the respiratory chain complex, in which they are involved in oxidative phosphorylation. It was also shown that SIRT3 proteins are involved in oxidative stress defense by protecting cells against reactive oxygen species (ROS). Moreover, during a caloric deficit, SIRT3 activate the key antioxidant mitochondrial enzyme—superoxide dismutase 2 (SOD2)—which increases mitochondria metabolism. In addition, SIRT3 activation prevents mitochondrial apoptosis (Sikora 2014).
Little is known about the function of other sirtuins in comparison with SIRT1 and SIRT3. SIRT2 play a role in homeostasis through the deacetylation of phosphoenolpyruvate carboxykinase, involved in gluconeogenesis. In contrast, the mitochondrial protein SIRT4 is responsible, among other, for the regulation of fatty acid β-oxidation in hepatocytes and myocytes. Interestingly, SITR3 and SITR4 play an opposite role in regulating the glutamate dehydrogenase (GDH), and fatty acid β-oxidation. SIRT5 protein causes deacetylation of carbamyl phosphate synthetase (CPS1), leading to more rapid detoxification of ammonia during the urea cycle. SIRT6 in turn is involved in stabilization and genomic DNA repair. They can also exert an anti-aging effect. Both of these functions may be related to the fact that SIRT6 are involved in the inhibition of retrotransposition of long interspersed nuclear elements 1 (LINE1) belonging to the non-LTR retrotransposons. LINE transposition damages cells and tissues causing their premature aging (Van Meter et al. 2014). Moreover, knockout mice, lacking the gene encoding SIRT6, were characterized by shorter life cycle, reduced insulin-like growth factor 1 (IGF-1) levels, the consequence of which was hypoglycemia (Mostoslavsky et al. 2006). In mouse cancer models, SIRT6 deficiency leads to increased tumor growth and aggressiveness (Sebastián et al. 2012). Although SIRT6 were localized in the nucleus, it was shown that they had an effect on mitochondrial homeostasis and affected the metabolism of cancer cells. In mitochondria, they enhanced intracellular respiration by inhibiting hypoxia-inducible factor-1 alpha (HIF1α), which in turn inhibited the transcription of key glycolytic enzymes. In tumor cells, they prevented the Warburg effect otherwise known as aerobic glycolysis (Zhong et al. 2010, 2013). SIRT7 activates RNA polymerase transcription; it is believed that it can also deacetylate p53 (Vakhrusheva et al. 2008; Houtkooper et al. 2012; Parihar et al. 2015). The study of Barber et al. (2012) indicates that SIRT7 is a nuclear regulator of both chromatin and mitochondria functions. It increases chromatin-dependent repression of genes involved in epigenetic stabilization of “oncogenic transformation”. It is assumed that SIRT7 may regulate mitochondrial homeostasis through deacetylation of GA-binding protein β1 (GABPβ1) (Fang et al. 2016). Tsai et al. (2014) showed that SIRT7 are involved in ribosome biogenesis and protein synthesis. Researchers indicated that SIRT7 regulates rDNA transcription and modulates PolIII functions. SIRT7 may also regulate transcription of PolIII through mTOR kinase and TFIIIC2 complex.
Regulation of activity
The expression of SIRT1 varies depending on physiological parameters. It is induced by lowering the energy state of the cell. For example, nutritional deficit enhances SIRT1 expression, while the high-fat diet reduces it. Within the promoter sequence of the SIRT1 gene, there are regions that bind transcription factors, such as forkhead box protein O1 (FOXO1), cAMP response element-binding (CREB) or carbohydrate response element-binding protein (CHREBP), suggesting that these factors regulate the transcription of SIRT1. At a higher level of expression regulation, microRNA (miRNA) affects the level of mRNA through degradation of pre-mRNA transcripts or translation inhibition. Less is known about the transcription control by other sirtuins. Studies in mice show that expression of STAT3 is activated by estrogen receptor α (ERRα), which is a nuclear receptor regulating the activity of mitochondria.
Regulation of SIRT1 activity at the level of post-translational modifications is the least understood. Studies on cell cultures, induced with ultraviolet rays or hydrogen peroxide, indicated SIRT1 inactivation, leading to cell death. Sirtuin activity is also controlled by the formation of protein complexes. AROS (active regulator of SIRT1) is a protein that regulates SIRT1 by forming complexes with it, leading to inhibition of its activity (Kim et al. 2007). LCD1 (histone methyltransferase, lysine-specific demethylase 1) also interacts with SIRT1 catalytic domains, thereby reducing the expression of genes encoding Notch receptor proteins (Mulligan et al. 2011).
It is also believed that the expression of SIRT6 may be dependent on SIRT1 through the formation of transcription complex (SIRT1-FOXO3A-NRF1) on the promoter of the gene encoding SIRT6. Furthermore, there are physical interactions between SIRT1 and SIRT7, which are important in the regulation of EMT-like process (the epithelial–mesenchymal transition) during tumor progression and metastasis (Fang et al. 2016).
The expression of sirtuins is also affected by the availability of the NADPH cofactor, as some of them require it for activation (Cantó et al. 2013).
Activation of SIRT1 can be enhanced by an increase in the level of NADH+. NADH precursors, i.e., nicotinamide riboside or nicotinamide mononucleotide (NMN) can also increase the activity of SIRT1. For example, in 22-month-old mice, weekly NMN supplementation caused the mitochondrial indicators to resemble indicators in young mice (Fang et al. 2016).
Naturally occurring compounds may also activate sirtuins. For example, in vitro studies demonstrated that several plant polyphenols, and resveratrol in particular, activate the expression of gene encoding the Sir2 protein, thereby leading to the deacetylation of p53 protein; treating the yeasts with this compound leads to a longer life cycle. Furthermore, studies in mice showed that resveratrol enhances SIRT1 activity in the mitochondria, thereby increasing their metabolism. Increased mitochondrial activity protects mice against obesity and strengthens their cold resistance. Mice treated with resveratrol, although fed a high-calorie diet, lived longer (Baur et al. 2006). In humans, resveratrol was also shown to increase mitochondrial activity and metabolic control (Timmers et al. 2011).
Sirtuins in plants
Sirtuins in plants are very poorly understood. Huang et al. (2007), among others, studied the function of genes coding for these proteins. They demonstrated that these genes are essential for protection against genomic instability and damage, guaranteeing cell growth in rice.
Plant genomes contain significantly less SIR2 homologs compared to other eukaryotes. The first plant genes encoding sirtuins were analyzed in rice (Oryza sativa) and were described as similar to genes encoding SIR2. Thus far, physiological function of plant SIR2 has not been determined. Huang et al. (2007) analyzed the function of genes similar to SIR2 in rice and they named them OsSRT1 and OsSRT2. OsSRT1 and other plant SRT1 homologs belong to class IV, while OsSRT2 to class II of genes related to SIR2. In plants, sirtuins belonging to class I and III have not been identified. OsSRT1 belongs to class IV of SIR2 proteins, likewise human SIRT6 and SIRT7; however, structurally it is more similar to SIRT6. Considering the fact that in humans, as mentioned above, SIRT6 proteins play a role in DNA damage repair and genomic instability, scientists speculate that the function of this protein in rice may be related (Mostoslavsky et al. 2006; Huang et al. 2007).
The plant SRT1 proteins exhibit a high degree of conservation. Only their N terminus is homologous to the animal sirtuins. Northern blotting analysis demonstrated that these proteins are active in a variety of tissues; however, a significant increase in the activity was observed in dividing cells, i.e., bud, seedling or inflorescence cells. Nuclear localization of these proteins has been also identified. Studies on the function of the protein encoded by the OsSRT1 gene using RNAi, decreasing its expression, showed an increase in the acetylation level and a decrease in H3K9 demethylation (lysine 9 in histone 3). Furthermore, the activation of mobile elements was found and increased expression of programmed cell death (PCD) marker genes, which was associated with the induction of apoptosis in the leaves and in consequence—premature aging. On the other hand, overexpression of OsSRT1 reduced H3K9 acetylation, thereby strengthening tolerance to oxidizing agents, which resulted in the enhanced tolerance to oxidative stress. Demethylation of H3K9 is associated with chromatin inactivation. The study results of Huang et al. (2007) suggest that OsSRT1 are involved in histone deacetylation necessary for transposon silencing. In summary, molecular analysis has shown that the function of OsSRT1 is associated with lysine deacetylation of histone-3 (H3), which is required for transcriptional repression of mobile elements and genes associated with apoptosis. Zhong et al. (2013) extended the study by Huang et al. (2007). The researchers decided to identify OsSRT1 binding sites. They did this by comparing genome-wide H3K9 acetylation profiles between wild-type and OsSRT1 down-regulated plants. Subsequently, they used the chromatin immunoprecipitation (ChIP) technique to investigate genomic binding sites. Analyses showed that H3K9Ac [acetyl-histone H3 (Lys 9)] was mostly associated with active genes in rice. Moreover, the OsSRT1 binding was found to be enriched over the gene bodies. It is also expected that OsSRT1 may be responsible for the regulation of genes related to the response induced by H3K9 deacetylation. In addition, the results indicate that OsSRT1 may be involved in metabolic regulation in plants and silencing transposable elements in rice. Chung et al. (2009) analyzed the subcellular location of histone deacetylases in rice. Scientists have compared the amino acid structure of these proteins and demonstrated that there was 39 % homology to the human OsSRT2 SiR3 (hSIR3) within the conserved domain. Furthermore, the scientists have localized this protein in the mitochondria, which suggests that OsSRT2 and hSIRT3 affect the metabolism of these organelles. Busconi et al. (2009) and Cucurachi et al. (2012) conducted research on sirtuins in Vitis vinifera L. Researchers have identified two genes encoding proteins similar to sirtuins in Vitis vinifera L. Both genes were localized in the nucleus. The first was mapped to chromosome 7, and nucleotide sequence of this gene was similar to mammalian SIRT4, which allowed to classify it to class II SIR2-like genes. The second gene was mapped to chromosome 19, and in terms of nucleotide sequence similarity, the genes correspond to class IVb of sirtuin-like genes. The results of subcellular localization studies indicated that the protein encoded by the VvSRT1 gene was also located in the nucleus and the protein encoded by VvSRT2 was most likely located in the stroma of chloroplasts and mitochondria. Comparative analysis of the coding sequences of these genes with other genes available in the database, i.e., in Arabidopsis thaliana and Oryza sativa, Ricinus communis, Populus trichocarpa or Brachypodium distachyon showed that their structure was highly conservative. Both proteins exhibited characteristics that allowed to classify their proteins into the SIR superfamily. Cucurachi et al. (2012) also performed an analysis of VvSRT1 and VvSRT2 gene expression, which demonstrated that VvSRT2 were characterized by higher expression changes in different tissues/organs, and particularly in the leaves when compared to VvSRT1, which expression level was substantially lower. It is possible that such a variable level of expression is caused by the different location of the proteins encoded by these genes, and thus their different functions. Probably, the photosynthetically active cells, i.e., leaf cells, contain many chloroplasts, which to function properly require a high level of sirtuins encoded by VvSRT2 genes, which in turn stabilize the chromatin structure and/or regulate the transcription of other chloroplast genes. These scientists supported their hypothesis by the research of Aquea et al. (2010), who observed VvSRT2 expression in the leaves, flowers and fruits. It was not detected only in the roots, which might indicate that a high level of the VvSRT2 gene expression was correlated with the activity of the photosynthetic cells.
The analysis of Arabidopsis thaliana genome sequence showed that only two SIR2 families (AtSRT1 and AtSRT2) are present in this species, while the phylogenetic analysis of SIR2 homologs showed that they also belong to two of the four classes of that family. AtSRT1 belongs to class IV and AtSRT2 to class II (Pandey et al. 2002; Hollender and Liu 2008). König et al. (2014) showed that AtSRT2 has the function of mitochondrial lysine deacetylase. The mitochondrial genome only contains genes encoding the AtSRT2, the presence of genes encoding AtSRT1 have not been detected. The AtSRT2 gene probably encodes seven isoforms of the RT2 protein. The study of König et al. (2014) showed the presence of two isoforms (AtSRT2A and AtSRT2B) that contained pre-sequences on the N-terminal corresponding to the mitochondrial SIR2; they corresponded to SRT2.1/SRT2.2/SRT2.3/SRT2.5 and SRT2.7 isoforms. Studies using C-terminal GFP fusion with AtSRT2 and Western blots showed mitochondrial localization of these isoforms. There was no evidence of nuclear localization of AtSRT2, although it was detected by Wang et al. (2010). The amino acid comparative analysis of AtSRT2 proteins with mammalian sirtuins revealed high similarity of these proteins to human SIRT4 (42 %), slightly lower to SIRT3 (31 %) and SIRT5 (25 %). It is believed that the AtSRT2 protein is associated with the inner mitochondrial membrane through protein–protein bonds. Probably, the activity of AtSRT2 as lysine deacetylase in mitochondria relates to specific proteins differing from those found in mammals. These include proteins associated with the inner mitochondrial membrane, i.e., ATP synthase and ATP/ADP carriers. Since the ATP synthesis is a regulator of energy metabolism in mitochondria, changes in the activity of this enzyme can in consequence alter the mitochondrial metabolism (König et al. 2014). Van Der Kelen et al. (2013) showed that reducing the expression of AtSIR2 decreased the response to salt stress. The analysis of intracellular interactions using tandem affinity purification (TAP) showed that AtSRT interact with chromatin remodeling factors in the nucleus, and are responsible for the deacetylation of histones, subunits of mitochondrial respiratory complex and cytoplasmic proteins. Wang et al. (2010) demonstrated that AtSRT2 is a negative regulator of the basal resistance in plants. AtSRT2 causes deacetylation within the promoter regions of PAD4 genes (phyto alexin deficient 4), EDSS (enhanced disease susceptibility) and SID2 (salicylic acid induction deficient), which results in their decreased expression, while it should be increased in response to pathogens, as they are classified among the pathogen-related (PR) genes.
Grozinger et al. (2001) analyzed the effect of various inhibitors on the expression of sirtuins and demonstrated that the inhibition of SRT2 expression in Arabidopsis through the use of sirtinol (a cell-permeable inhibitor of sirtuin NAD+-dependent deacetylases) changed the structure of the leaves, and also led to a distortion of plant symmetry axis. In addition, vascular system formation in seedlings was disturbed. It was suspected that SRT2-like proteins are responsible for the growth and development of plants (Grozinger et al. 2001). Plants treated with sirtinol were characterized by thick, short hypocotyls and the lack of major roots, as well as abnormal cotyledon venation. Similar disorders are observed in the plants with impaired auxin transport. These authors hypothesized that sirtuins might control the synthesis of auxins or proteins involved in auxin transport or they could regulate the cellular response to auxin action (Grozinger et al. 2001).
Cigliano et al. (2013), and later Zhao et al. (2015), analyzed histone deacetylases in tomato (Solanum lycopersicum). Of the gene pool belonging to the HDAC family, they found two proteins similar to SIR2—SlSRT1 and SlSRT2. SRT2 was found in the nucleus (Zhao et al. 2015). Cigliano et al. (2013) analyzed the expression of both genes. They observed increased expression of the SlSRT1 gene in the buds and 1-cm fruits and SlSRT2 in flowers and fruits. These authors by analyzing the results deduced that SIRT1 proteins were probably involved in the development of fruits, while SlSRT2 in their subsequent maturation and also in gametogenesis.
In addition, the presence of genes encoding proteins similar to sirtuins was found in the genomes of maize (Chandler et al. 2004), wheat, rapeseed and lettuce, sorghum or poplar. However, besides the sequences of those genes deposited in the National Center for Biotechnology (NCBI), further information is still missing about the products of these genes, their function, location, etc., which leaves an open space for researchers that investigate this issue (Table 2).
Summary
Considering the above reports, and first of all, the fact that animal sirtuins may affect the processes of aging, rejuvenation and regeneration, research expanding the current knowledge on this subject is very important. All the more that they are a promising molecular target for the treatment of many diseases associated with old age. Increasingly, more scientific reports indicate that the skillful control of their activity using chemical activators, i.e., resveratrol or proper diet can delay aging and prolong the life span. At present, compounds that stimulate the activation of sirtuins are widely used in beauty industry. Furthermore, it is essential to expand the knowledge about plant sirtuins, which probably affect the growth and development of plants as well as protect against genomic instability. With the increasing availability of molecular techniques and bioinformatic tools, the knowledge of these yet still vaguely known enzymes expands. More and more reports indicate that the function of plant sirtuins is similar to those found in animals. In addition to these listed above, they are also believed to play a role in controlling the synthesis of such an important phytohormone for plants such as auxin, as well as in defensive responses against the virulent plant pathogen—P. syringae. It is also thought that they are involved in the ripening of the fruits and gametogenesis. Moreover, their function was also correlated with photosynthetic activity and aging of leaves. The increasing knowledge about the plant sirtuins provides more and more opportunities to discover plant sirtuin modulators. This knowledge may be also found useful for breeding purposes or gardening. It is possible that further research, especially concerning the manipulation of sirtuin gene expression and modulation of the activity of these enzymes will allow scientists to affect the rate of maturation of plants, which in turn would create both large economic and social benefits.
Author contribution statement
Main author review: Sirtuins—not only animal proteins. Izabela Szućko—study conception and design, collection of data, and writing the manuscript.
References
Aquea F, Timmermann T, Arce-Johnson P (2010) Analysis of histone acetyltransferase and deacetylase families of Vitis vinifera. Plant Physiol Biochem 48:194–199
Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, Struhl K, Garcia BA, Gozani O, Li W, Chua KF (2012) SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487:114–118
Baur JA, Pearson KJ, Price NL, Jamieson H, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair D (2006) Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444(7117):337–342
Busconi M, Reggi S, Fogher C, Bavaresco L (2009) Evidence of a sirtuin gene family in grapevine (Vitis vinifera L.). Plant Physiol Biochem 47(7):650–652
Cantó C, Gerhart-hines Z, Feige JN, Lagouge M, Milne JC, Elliott PJ, Puigserver P, Auwerx J (2013) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458(7241):1056–1060
Chandler VL, Kaeppler SM, Kaeppler HF, Cone KC (2004) Sequences from the Plant Chromatin Consortium NSF (Plant Genome project 9975930) (unpublished)
Chung PJ, Kim YS, Parka S-H, Nahm BH, Kim J-K (2009) Subcellular localization of rice histone deacetylases in organelles. FEBS Lett 583:2249–2254
Cigliano R, Sanseverino W, Cremona G, Ercolano MR, Conicella C, Consiglio FM (2013) Genome-wide analysis of histone modifiers in tomato: gaining an insight into their developmental roles. BMC Genomics 14:57
Cucurachi M, Busconi M, Morreale G, Zanetti A, Bavaresco L, Fogher C (2012) Characterization and differential expression analysis of complete coding sequences of Vitis vinifera L. sirtuin genes. Plan Physiol Biochem 54:123–132
Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA (2014) Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 157(4):882–996
Fang EF, Scheibye-Knudsen M, Chua KF, Mattson MP, Croteau DL, Bohr VA (2016) Nuclear DNA damage signalling to mitochondria in ageing. Nat Rev Mol Cell 17(5):308–321
Frye R (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273(2):793–798
Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL (2001) Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem 76(42):38837–38843
Hollender C, Liu Z (2008) Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol 50(7):875–885
Houtkooper RH, Pirinen E, Auwerx J (2012) Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13(4):225–238
Huang L, SunQ Qin F, Li C, Zhao Y, Zhou D-H (2007) Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol 144(3):1508–1519
Kim EJ, Kho JH, Kang MR, Um SJ (2007) Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell 28(2):277–290
König A-C, Hartl M, Pham PA, Laxa M, Boersema PJ, Orwat A, Kalitventseva I, Plöchinger M, Braun H-P, Leister D, Mann M, Wachter A, Fernie AR, Finkemeier I (2014) The Arabidopsis class II sirtuin is a lysine deacetylase and interacts with mitochondrial energy metabolism. Plant Physiol 164(3):1401–1414
Kupis W, Pałyga J, Tomal E, Niewiadomska E (2016) The role of sirtuins in cellular homeostais. J Physiol Biochem. doi:10.1007/s13105-016-0492-6
Li K, Casta A, Wang R, Lozad E, Fan W, Kane S, Ge Q, Gu W, Orren D, Luo J (2008) Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Biol Chem 283(12):7590–7598
Moniot S, Weyand M, Steegborn C (2012) Structures, substrates, and regulators of mammalian sirtuins—opportunities and challenges for drug development. Front Pharmacol 3(16):1–5
Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Gellon MR, Liu L, Mostoslavsky P, Franco G, Murphy S, Mills MM, Katel KD, Hsu P, Hong JT, Ford AL, Cheng E, Kennedy HL, Nunez C, Bronson N, Frendewey R, Auerbach D, Valenzuela W, Karow D, Hottiger M, Hursting MO, Barrett S, Guarente JC, Mulligan L, Demple R, Yancopoulos B, Alt GD (2006) Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124(2):315–329
Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, Mostoslavsky R, Gygi SP, Gill G, Dyson NJ, Näär AM (2011) A SIRT1-LSD1 corepressor complex regulates notch target gene expression and development. Mol Cell 42(5):689–699
Pandey R, Müller A, Napoli C, Selinger D, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen R (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30(23):5036–5055
Parihar P, Solanki I, Mansuri M, Parihar MS (2015) Mitochondrial sirtuins: emerging roles in metabolic regulations, energy homeostasis and diseases. Exp Gerontol 61:130–141
Sebastián C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, Cosentino C, Greenson JK, MacDonald AI, McGlynn L, Maxwell F, Edwards J, Giacosa S, Guccione E, Weissleder R, Bernstein BE, Regev A, Shiels PG, Lombard DB, Mostoslavsky R (2012) The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151(6):1185–1199
Shoba B, Lwin ZM, Ling LS, Bay BH, Yip GW, Kumar SD (2009) Function of sirtuins in biological tissues. Anat Rec 292(4):536–543
Siedlecka K, Bogusławski W (2005) Sirtuins—longevity enzymes? Gerontol Pol 13(3):147–152 (in Pol.)
Sikora E (2014) Aging and longevity. Postępy Biochemii 60(2):125–137 (in Pol.)
Timmers S, Konings E, Bilet L, Houtkooper RH, Van De Weijer T, Goossens GH, Hoeks J, Van Der Krieken S, Ryu D, Kersten S, Moonen-Kornips E, Hesselink MKC, Kunz I, Schrauwen-Hinderling VB, Blaak EE, Auwerx J, Schrauwen P (2011) Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab 14(5):612–622
Tissenbaum H, Guarente L (2001) Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410(6825):227–230
Tsai Y-C, Greco TM, Cristea M (2014) Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. MCP 13(1):73–83
Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E (2008) Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res 102(6):703–710
Van Der Kelen K, Hannah M, Gossele V, Metzlaff M, Van Breusegem F (2013) A functional study of Arabidopsis thaliana sirtuins. In: International plant and animal genome conference XXI
Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V (2014) SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun 5:5011
Wang C, Gao F, Wu J, Dai J, Wei C, Li Y (2010) Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol 51(8):1291–1299
Whitaker R, Faulkner S, Miyokawa R, Burhenn L, Henriksen M, Wood JG, Helfand SL (2013) Increased expression of Drosophila sir2 extends life span in a dose dependent manner. Aging (Albany. NY) 5(9):682–691
Yamamori T, DeRicco J, Naqvi A, Hoffman TA, Mattagajasingh I, Kasuno K, Jung S-B, Kim C-S, Irani K (2010) SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res 38(3):832–845
Zhao L, Lu J, Zhang J, Wu P-Y, Yang S, Wu K (2015) Identification and characterization of histone deacetylases in tomato (Solanum lycopersicum). Front Plant Sci 5(760):1–9
Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R (2010) The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140(2):280–293
Zhong X, Zhang H, Zhao Y, Sun Q, Hu Y, Peng H, Zhou D-X (2013) The rice NAD+-dependent histone deacetylase OsSRT1 targets preferentially to stress- and metabolism-related genes and transposable elements. PLoS One 8(6):1–8
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by AK Kononowicz.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Szućko, I. Sirtuins: not only animal proteins. Acta Physiol Plant 38, 237 (2016). https://doi.org/10.1007/s11738-016-2255-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-016-2255-y