Abstract
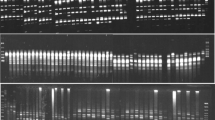
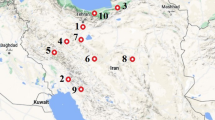
Information on population genetic structure and crop genetic diversity is important for genetically improving crop species and conserving threatened species. The PAL gene sequence is part of a multigene family that can be utilized to design DNA marker systems for genetic diversity and population structure investigation. In the current study, genetic diversity and population structure of 100 accessions of wild Pistacia species were investigated with 78 PAL markers. A protocol for using PAL sequences as DNA markers was developed. A total of 313 PAL loci were recognized, showing 100% polymorphism for PAL markers. The PAL markers produced relatively more observed and effective alleles in Pistacia falcata and Pistacia atlantica, with a higher Shannon’s information index and expected heterozygosity in P. atlantica, Pistacia vera and Pistacia mutica. Pairwise assessment of Nei’s genetic distance and genetic identity between populations revealed a close association between geographically isolated populations of Pistacia khinjuk and Pistacia chinensis. The accessions of wild Pistacia species had more genetic relationship among studied groups of species. Analysis of molecular variance indicated 19% among-population variation and 81% within-population variation for the PAL gene based DNA marker. Population structure analysis based on PAL revealed four groups with high genetic admixture among populations. The results establish PAL markers as a functional DNA marker system and provide important genetic information about accessions from wild populations of Pistacia species.

Similar content being viewed by others
Abbreviations
- PAL:
-
Phenylalanine ammonia-lyase
- PCRs:
-
Polymerase chain reactions
- PAGE:
-
Polyacrylamide gel electrophoresis
- PIC:
-
Polymorphism information content
- RP:
-
Resolving power
- SI:
-
Shannon’s information index
- AMOVA:
-
Analysis of molecular variance
- CCC:
-
Cophenetic correlation coefficient
- UPGMA:
-
Unweighted pair group method arithmetic
- PCA:
-
Principal component analysis
- PCoA:
-
Principal coordinate analysis
- NTSYS:
-
Numerical taxonomy multivariate analysis system
- MCMC:
-
Markov chain Monte Carlo
- SCoT:
-
Start codon targeted
- IRAP:
-
Inter-retrotransposon amplified polymorphism
- MI:
-
Marker index
- PP:
-
Percentage polymorphism
- UTR:
-
Untranslated region
References
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186. https://doi.org/10.1139/g93-024
Bagal UR, Leebens-Mack JH, Lorenz WW, Dean JF (2012) The phenylalanine ammonia lyase (PAL) gene family shows a gymnosperm-specific lineage. BMC Genom 13(Suppl. 3):S1
Basha SA, Chatterjee SC (2007) Activation of phenylalanine ammonia lyase contributes to non-host resistance in Triticum aestivum against Sclerotinia sclerotiorum. Indian Phytopathol 60:442–449
Campos R, Nonogaki H, Suslow T, Saltveit ME (2004) Isolation and characterization of a wound inducible phenylalanine ammonia-lyase gene (LsPAL1) from Romaine lettuce leaves. Physiol Plant 121:429–438. https://doi.org/10.1111/j.1399-3054.2004.00336.x
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Chang A, Lim MH, Lee SW, Robb EJ, Nazar RN (2008) Tomato phenylalanine ammonia-lyase gene family, highly redundant but strongly underutilized. J Biol Chem 283:33591–33601. https://doi.org/10.1074/jbc.M804428200
Creste S, Neto AT, Figueira A (2001) Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Plant Mol Biol Rep 19:299–306. https://doi.org/10.1007/BF02772828
Cui Y, Magill J, Frederiksen R, Magill C (1996) Chalcone synthase and phenylalanine ammonia lyase mRNA following exposure of sorghum seedlings to three fungal pathogens. Physiol Mol Plant Pathol 49:187–199. https://doi.org/10.1006/pmpp.1996.0048
Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097
Dong CJ, Shang QM (2013) Genome-wide characterization of phenylalanine ammonia-lyase gene family in watermelon (Citrullus lanatus). Planta 238:35–49. https://doi.org/10.1007/s00425-013-1869-1
Duroux L, Welinder KG (2003) The peroxidase gene family in plants: a phylogenetic overview. J Mol Evol 57:397–407. https://doi.org/10.1007/s00239-003-2489-3
Esmail-pour A (2001) Distribution, use and conservation of pistachio in Iran. In: Padulosi S, Hadj-Hassan A (eds), Towards a comprehensive documentation and use of Pistacia genetic diversity in central and West Asia, North Africa and Europe, 16–26. Report of the IPGRI Workshop, 14–17 Dec 1998, Ibrid, Jordan. International Plant Genetic Resources Institute (IPGRI), Rome, Italy
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null allele. Mol Ecol 7:574–578. https://doi.org/10.1111/j.1471-8286.2007.01758.x
FAO (2006) FAOSTAT database. www.fao.org/faostat/
Fraser CM, Chapple C (2011) The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book 9:e152
Fukasawa-Akada T, Kung SD, Watson JC (1996) Phenylalanine ammonia-lyase gene structure, expression, and evolution in Nicotiana. Plant Mol Biol 30:711–722
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930. https://doi.org/10.1016/j.plaphy.2010.08.016
Gonzalez AM, Marcel TC, Kohutova Z, Stam P, van der Linden CG, Niks RE (2010) Peroxidase profiling reveals genetic linkage between peroxidase gene clusters and basal host and nonhost resistance to rusts and mildew in barley. PLoS ONE 5(8):e10495. https://doi.org/10.1371/journal.pone.0010495
Gulsen O, Shearman RC, Heng-Moss TM, Mutlu N, Lee DJ, Sarath G (2007) Peroxidase gene polymorphism in buffalograss and other grasses. Crop Sci 47:767–774. https://doi.org/10.2135/cropsci2006.07.0496
Gulsen O, Kaymak S, Ozongun S, Uzun A (2010) Genetic analysis of Turkish apple germplasm using peroxidase gene-based markers. Sci Hortic 125:368–373
Hamberger B, Ellis M, Friedmann M, de Azevedo Sousa C, Barbazuk B, Douglas C (2007) Genome-wide analyses of phenylpropanoid- related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene families. Can J Bot 85:1182–1201
Herrero J, Esteban-Carrasco A, Zapata JM (2013) Looking for Arabidopsis thaliana peroxidases involved in lignin biosynthesis. Plant Physiol Biochem 67:77–86. https://doi.org/10.1016/j.plaphy.2013.02.019
Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H (2001) A large family of class III plant peroxidase. Plant Cell Physiol 42:462–468. https://doi.org/10.1093/pcp/pce061
Iranjo P, Nabati Ahmadi D, Sorkheh K, Rajabi Memeari H, Ercisli S (2016) Genetic diversity and phylogenetic relationships between and within wild Pistacia species populations and implications for its conservation. J For Res 27:685–697. https://doi.org/10.1007/s11676-015-0098-9
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bulletin de la Société Vaudoise des Sciences Naturelles 44:223–270
Jeong MJ, Choi BS, Bae DW, Shin SC, Park SU, Lim HS, Kim J, Kim JB, Cho BK, Bae H (2012) Differential expression of kenaf phenylalanine ammonia-lyase (PAL) ortholog during developmental stages and in response to abiotic stresses. Plant Omics J 5:392–399
Jolliffe IT (2002) Principal component analysis, 2nd edn. Springer, New York
Jones DH (1984) Phenylalanine ammonia-lyase: regulation of its induction, and its role in plant development. Phytochemistry 23:1349–1359. https://doi.org/10.1016/S0031-9422(00)80465-3
Kao YY, Harding SA, Tsai CJ (2002) Differential expression of two distinct phenylalanine ammonia-lyase genes in condensed tannin-accumulating and lignifying cells of quaking aspen. Plant Physiol 130:796–807
Khan MSY, Bano S, Javed K, Mueed MA (2006) A comprehensive review of the chemistry and pharmacology of Corchorus species—A source of cardiac glycosides, triterpenoids, ionones, flavonoids, coumarins, steroids and some other compounds. J Sci Ind Res 65:283–298
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49:725–738
Legendre P, Legendre L (1998) Numerical ecology, 2nd English edn. Elsevier, Amsterdam
Lewontin RC (1972) The apportionment of human diversity. In: Dobzhansky T, Hecht M, Steere WC (eds) Springer book archive, evolutionary biology, pp 381–398. 10.1007/978-1-4684-9063-3_14
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
McDermott JM, McDonald BA (1993) Gene flow in plant pathosystems. Annu Rev Phytopathol 31:353–373. https://doi.org/10.1146/annurev.py.31.090193.002033
Milligan BG, Leebens-Mack J, Strand AE (1994) Conservation genetics: beyond the maintenance of marker diversity. Mol Ecol 12:844–855
Mittler R, Vanderauwera S, Gollery M, Breusegem FW (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498. https://doi.org/10.1016/j.tplants.2004.08.009
Mizutani M, Ohta D, Sato R (1997) Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in plant. Plant Physiol 113:755–763
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci (USA) 70:3321–3323
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Olsen KM, Lea US, Slimestad R, Verheul M, Lillo C (2008) Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J Plant Physiol 165:1491–1499
Passardi F, Cosio C, Penel C, Dunand C (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24:255–265. https://doi.org/10.1007/s00299-005-0972-6
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Prevost A, Wilkinson MJ (1999) A new system of comprising PCR primers applied to ISSR fingerprinting of potato accessions. Theor Appl Genet 98:107–112. https://doi.org/10.1007/s001220051046
Pritchard JK, Stevens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen XQ, Falush D (2010) Documentation for structure software: Version 2.3. http://pritch.bsd.uchicago.edu/software/structure_v.2.3.1/documentation.pdf. Accessed 15 Sept 2012
Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133:1051–1071
Ramakrishnan M, Antony Ceasar S, Duraipandiyan V, Al-Dhabi NA, Ignacimuthu S (2016) Using molecular markers to assess the genetic diversity and population structure of finger millet (Eleusine coracana (L.) Gaertn.) from various geographical regions. Genet Resour Crop Evol 63:361–376. https://doi.org/10.1007/s10722-015-0255-1
Rawal HC, Singh NK, Sharma TR (2013) Conservation, divergence, and genome-wide distribution of PAL and POX A gene families in plants. Int J Genomics. Article ID 678969, p 10. http://dx.doi.org/10.1155/2013/678969
Razavi S (2006) Pistachio production, Iran vs. the world. Acta Hortic 726:689–694
Reichert AI, He XZ, Dixon RA (2009) Phenylalanine ammonia-lyase (PAL) from tobacco (Nicotiana tabacum): characterization of the four tobacco PAL genes and active heterotetrameric enzymes. Biochem J 424:233–242. https://doi.org/10.1042/BJ20090620
Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, Van Driessche G, Van Beeumen J, Messens E, Boerjan W (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16:2749–2771
Rohlf M (1998) NTSYSpc. Numerical taxonomy and multivariate analysis system. Version 2.02i. Department of Ecology and Evolution. State University of New York
Satya P, Banerjee R, Biswas C, Karan M, Ghosh S, Ali N (2014) Genetic analysis of population structure using peroxidase gene and phenylalanine ammonia-lyase gene-based DNA markers: a case study in Jute (Corchorus spp.). Crop Sci 54:1609–1620. https://doi.org/10.2135/cropsci2013.08.0518
Schopfer P (2001) Hydroxyl radical-induced cell-wall loosening in vitro and in vivo: implications for the control of elongation growth. Plant J 28:679–688. https://doi.org/10.1046/j.1365-313x.2001.01187.x
Shang QM, Li L, Dong CJ (2012) Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L. Planta 236:1093–1105
Sorkheh K, Shiran B, Gradziel TM, Epperson BK, Martinez-Gomez P, Asadi E (2007) Amplified fragment length polymorphism as a tool for molecular characterization of almond germplasm: genetic diversity among cultivated genotypes and related wild species of almond, and its relationships with agronomic traits. Euphytica 156:237–344
Sorkheh K, Amirbakhtiar N, Ercislie S (2016) Potential start codon targeted (SCoT) and inter-retrotransposon amplified polymorphism (IRAP) markers for evaluation of genetic diversity and conservation of wild Pistacia species population. Bioch Genet 54:368–387. https://doi.org/10.1007/s10528-016-9725-1
Yeh FC, Yang R, Boyle TJ, Ye Z, Xiyan JM (2000) PopGene32, microsoft windows-based freeware for population genetic analysis. Version 1.32. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton
Zhang ZG, Zhang Q, Wu JX, Zheng X, Zheng S, Sun XH, Qiu QS, Lu TG (2013) Gene knockout study reveals that cytosolic Ascorbate Peroxidase 2(OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 8(2):E57472. https://doi.org/10.1371/journal.pone.0057472
Zohary D (1996) The genus Pistacia L. In: Padulosi S, Caruso T, Barone E (eds) Taxonomy, distribution, conservation and uses of Pistacia genetic resources. IPGRI, Palermo, Italy, pp 1–11
Acknowledgements
We thank the Iranian Pistachio Research Institute, Rafsanjan, Iran for providing some samples of the accessions of wild Pistacia species. We also thank two anonymous reviewers for constructive suggestions that greatly helped to improve the manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to this work.
Corresponding author
Additional information
Project funding: This study was supported by Shahid Chamran University of Ahvaz Fund (SHCUF) under Project No. SHCH_AGF_Grant 1394.
The online version is available at http://www.springerlink.com
Corresponding editor: Tao Xu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mirzavand, S., Sorkheh, K. & Siahpoosh, M.R. Genetic diversity and population structure analysis of Pistacia species revealed by phenylalanine ammonia-lyase gene markers and implications for conservation. J. For. Res. 29, 991–1001 (2018). https://doi.org/10.1007/s11676-017-0508-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-017-0508-2