Abstract
Purpose
The confusion of MRI sequence names could be solved if MR images were automatically identified after image data acquisition. We revealed the ability of deep learning to classify head MRI sequences.
Materials and methods
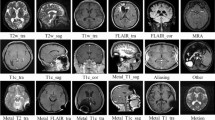
Seventy-eight patients with mild cognitive impairment (MCI) having apparently normal head MR images and 78 intracranial hemorrhage (ICH) patients with morphologically deformed head MR images were enrolled. Six imaging protocols were selected to be performed: T2-weighted imaging, fluid attenuated inversion recovery imaging, T2-star-weighted imaging, diffusion-weighted imaging, apparent diffusion coefficient mapping, and source images of time-of-flight magnetic resonance angiography. The proximal first image slices and middle image slices having ambiguous and distinctive contrast patterns, respectively, were classified by two deep learning imaging classifiers, AlexNet and GoogLeNet.
Results
AlexNet had accuracies of 73.3%, 73.6%, 73.1%, and 60.7% in the middle slices of MCI group, middle slices of ICH group, first slices of MCI group, and first slices of ICH group, while GoogLeNet had accuracies of 100%, 98.1%, 93.1%, and 94.8%, respectively. AlexNet significantly had lower classification ability than GoogLeNet for all datasets.
Conclusions
GoogLeNet could judge the types of head MRI sequences with a small amount of training data, irrespective of morphological or contrast conditions.



Similar content being viewed by others
References
Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Proceedings of the 25th international conference on neural information processing systems, vol. 1. Lake Tahoe, Nevada. 2999257: Curran Associates Inc.; 2012. p. 1097–105.
Szegedy C, Liu W, Jia Y, Sermanet P, Reed SE, Anguelov D, et al. Going deeper with convolutions. In: 2015 IEEE conference on computer vision and pattern recognition (CVPR). 2015; p. 1–9.
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016;35(5):1299–312.
Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–98.
Cheng PM, Malhi HS. Transfer learning with convolutional neural networks for classification of abdominal ultrasound images. J Digital Imaging. 2017;30(2):234–43.
Obuchowski NA. Receiver operating characteristic curves and their use in radiology. Radiology. 2003;229(1):3–8.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77.
Wang S, Summers RM. Machine learning and radiology. Med Image Anal. 2012;16(5):933–51.
McCulloch WS, Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull Math Biophys. 1943;5(4):115–33.
Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–82.
Shi B, Grimm LJ, Mazurowski MA, Baker JA, Marks JR, King LM, et al. Prediction of occult invasive disease in ductal carcinoma in situ using deep learning features. J Am Coll Radiol. 2018;15:527–34.
Masood A, Sheng B, Li P, Hou X, Wei X, Qin J, et al. Computer-assisted decision support system in pulmonary cancer detection and stage classification on CT images. J Biomed Inform. 2018;79:117–28.
Santini G, Latta DD, Martini N, Valvano G, Gori A, Ripoli A, et al., editors. An automatic deep learning approach for coronary artery calcium segmentation. Singapore: Springer; 2018.
Korfiatis P, Kline TL, Erickson BJ. Automated segmentation of hyperintense regions in FLAIR MRI using deep learning. Tomography. 2016;2(4):334–40.
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–52.
Hastie T, Tibshirani R, Friedman J. 7.11.1 Example (Continued). In: The elements of statistical learning: data mining, inference, and prediction. 2nd ed. New York, NY: Springer. 2009; p. 252–4.
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research from Japan society for the promotion of science (16K10333).
Funding
This work was supported in part by Grants-in-Aid for Scientific Research from Japan society for the promotion of science (16K10333).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Ethical statement
This study was approved by our hospital's institutional review board, which waived the need for written informed consent from the patients in light of the retrospective study design.
About this article
Cite this article
Noguchi, T., Higa, D., Asada, T. et al. Artificial intelligence using neural network architecture for radiology (AINNAR): classification of MR imaging sequences. Jpn J Radiol 36, 691–697 (2018). https://doi.org/10.1007/s11604-018-0779-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11604-018-0779-3