Abstract
The systematic position of two strains of a yeast-like fungus isolated from plant remnants on the Kharg Island in the Persian Gulf of Iran is evaluated using morphological, physiological and phylogenetic analyses. In culture, this fungus produced cylindrical cells that reproduced by polar budding on short stalks. Production of ballistoconidia and blastospores was observed. The carbon source assimilation spectrum was broad, but fermentation ability was absent. Phylogenetic analyses of the nuclear SSU, LSU (D1/D2 domain), and ITS rDNA revealed that this fungus represents a new lineage in the Urocystidales of the subphylum Ustilaginomycotina. Based on the comparison of phenotypic characters, physiology, and DNA sequences, a new genus and species Fereydounia khargensis (IBRC-M 30116T = CBS 13305T) is described for this fungus and accommodated in the novel family Fereydouniaceae. This is the first report of anamorphic saprobic fungus residing in the Urocystidales, stressing the remarkable evolutionary diversity in the subphylum Ustilaginomycotina.
Similar content being viewed by others
Introduction
The subphylum Ustilaginomycotina includes diverse basidiomycete fungi that are currently assembled in four main lineages accepted as classes, the Exobasidiomycetes, Ustilaginomycetes (Begerow et al. 2014), Malasseziomycetes, and Moniliellomycetes (Wang et al. 2014). The members of Ustilaginomycotina are unified by physiological, ultrastructural, and molecular characters, having a distinctive cell wall carbohydrate composition with a dominance of glucose and the absence of xylose, septal pores (in most species) enclosed at both sides by membrane caps, and zones of host–parasite interaction with fungal deposits resulting from exocytosis of primary interactive vesicles (Bauer et al. 1997, 2001b, 2006; Begerow et al. 2007, 2014); as well as forming a monophyletic group in broad-sampling fungal phylogenies (Hibbett et al. 2007; Matheny et al. 2007; Begerow et al. 2007, 2014). The subphylum Ustilaginomycotina comprises species with diverse life strategies and organization, including plant parasitic teliosporic species (classical smut fungi), plant parasitic nonteliosporic species, saprobic yeasts lacking or with a thus far unknown parasitic phase, and zoophilic lipophilic yeasts (Bauer et al. 2001b; Begerow et al. 2014).
Most ustilaginomycotinous species are dimorphic, i.e., their life cycle involves a saprobic yeast phase and a plant parasitic dikaryophase (Bauer et al. 2001b; Sampaio 2004; Begerow et al. 2014), but from many species, only a saprobic yeast phase is known: either these species lost the ability to develop a plant parasitic phase or this phase is currently unknown. These species are unevenly distributed through the ustilaginomycotinous tree of life, being known in seven of the 11 currently recognized orders of this fungal subphylum. Plant parasitic species are lacking in the Malasseziales and Moniliellales, which include only anamorphic yeast species classified in the genera Malassezia Baill. and Moniliella Stolk & Dakin, respectively (Begerow et al. 2000; Sampaio 2004; Wang et al. 2014). The order Ustilaginales contains a huge number of plant parasitic teliosporic species, mostly members of Poaceae and Cyperaceae, and comprises several anamorphic species classified in the genera Pseudozyma Bandoni emend. Boekhout, typified by P. prolifica Bandoni, and Farysizyma Á. Fonseca, typified by F. itapuensis Landell & P. Valente. Pseudozyma species are polyphyletic (mostly) within the ustilaginacean lineage, assigned to the so-called Ustilago-Sporisorium-Macalpinomyces complex (Begerow et al. 2000; Sampaio 2004; Stoll et al. 2005) that is currently split into several distinct genera (McTaggart et al. 2012). Farysizyma accommodates asexual members of the plant parasitic teliosporic genus Farysia Racib. (Inácio et al. 2008). In the spirit of the current International Code of Nomenclature for algae, fungi, and plants (ICN), in the future, probably all Pseudozyma and Farysizyma species will have to be merged with the respective teliosporic genera, since apparently all of them are only asexual morphs of known sexual genera and do not represent distinct lineages deserving a generic name.
The order Entylomatales, with one sexual genus, Entyloma de Bary, includes the anamorphic genus Tilletiopsis Derx. The type species T. washingtonensis Nyland and some related species form a distinct lineage (Begerow et al. 2000; Sampaio 2004; Boekhout et al. 2006) and probably deserve generic status. Several other Tilletiopsis-like species are apparently only asexual morphs of either the teliosporic genus Entyloma (Boekhout et al. 2006), or have affinity to the teliosporic genera Jamesdicksonia Thirum., Pavgi & Payak, Gjaerumia R. Bauer, M. Lutz & Oberw., and Phragmotaenium R. Bauer, Begerow, A. Nagler & Oberw. in the Georgefischeriales (Bauer et al. 2001a, 2005; Boekhout et al. 2006). Two species (Tilletiopsis albescens Gokhale, T. pallescens Gokhale) cannot be assigned to any major group of the Ustilaginomycotina (Begerow et al. 2000, 2007; Boekhout et al. 2006).
The orders Exobasidiales and Microstromatales include diverse assemblages of sexual and asexual species. In the former order, anamorphic species are classified in two asexual genera: Acaromyces Boekhout, Scorzetti, Gerson & Sztejnb., typified by A. ingoldii Boekhout, Scorzetti, Gerson & Sztejnb., and Meira Boekhout, Scorzetti, Gerson & Sztejnb., typified by M. geulakonigii Boekhout, Scorzetti, Gerson & Sztejnb. (Boekhout et al. 2003). In the latter order, anamorphic species are assigned to the following three asexual genera: Jaminaea Sipiczki & Kajdacsi, typified by J. angkorensis Sipiczki & Kajdacsi, Sympodiomycopsis Sugiy., Tokuoka & Komag., typified by S. paphiopedili Sugiy., Tokuoka & Komag., and Quambalaria J.A. Simpson, typified by Q. pitereka (J. Walker & Bertus) J.A. Simpson (Sugiyama et al. 1991; Simpson 2000; de Beer et al. 2006; Sipiczki and Kajdacsi 2009), while some other species are apparently only asexual morphs of known sexual genera (de Beer et al. 2006). To date, yeast-like saprobic species are unknown in the orders Ceraceosorales, Doassansiales, Tilletiales, and Urocystidales.
The plant parasitic teliosporic members of the subphylum Ustilaginomycotina, i.e., classical smut fungi, were recently studied in Iran by Vánky and Abbasi (2013), resulting in a monographic publication that includes descriptions, illustrations, host plants and the geographic distribution of 15 genera with 96 species occurring in this country (and one genus with three species of false smuts classified in the pucciniomycotinous order Microbotryales). Six of these species [Anthracoidea songorica Vánky, Entyloma majewskii Vánky & M. Lutz, Tilletia rostrariae Vánky & Ershad, Tranzscheliella iranica (Syd.) Vánky, Urocystis behboudii (Esfand.) Vánky, and Urocystis phalaridis Vánky] are currently known only from Iran (Vánky and Abbasi 2013). One additional genus with one species was added afterwards (Vasighzadeh et al. 2014), but still, the potential diversity of classical smut fungi in Iran is probably much higher. In contrast to the plant parasitic teliosporic species, the information on most of the remaining members of the subphylum Ustilaginomycotina occurring in this country is principally unavailable; the exceptions are studies on Pseudozyma fusiformata (Buhagiar) Boekhout as a biocontrol agent against Aspergillus flavus Link (Afsah-Hejri 2013), and studies on Malassezia species causing human skin diseases in Iran (Moniri et al. 2009; Shokohi et al. 2009; Hedayati et al. 2010; Zarei-Mahmoudabadi et al. 2013).
In the course of biodiversity research focusing on yeast species occurring in native environments of Iran, two strains of a yeast-like fungus have been isolated from plant remnants on Kharg Island in the Persian Gulf. The morphological, physiological and molecular analyses revealed the affinity of these strains to plant parasitic teliosporic species (classical smut fungi) classified in the order Urocystidales. This study aims at resolving the identity of these two strains and the phylogenetic placement within the Ustilaginomycotina.
Materials and methods
Isolation, morphology, physiology, and carbohydrate analysis
Residual plant blocks were collected aseptically and macerated in 90 ml saline. Dilutions from this sample were directly plated on Rose Bengal agar (Himedia, Mumbai, India) supplemented with 0.1 g/l chloramphenicol, followed by incubation at 28 °C for 48 h. Representatives of different morphological types of yeast colonies were isolated and purified by streaking on yeast peptone glucose (YPG) agar medium at 28 °C. Two isolates that formed similar colonies were selected for further examination. To assess colony morphology and sizes, the isolates were grown on YPG agar, 2 % malt extract agar (MEA, Merck, Germany), dichloran 18 % glycerol agar (DG18, Merck, Germany) and malt yeast 40 % sucrose agar (M40Y) (Nguyen et al. 2013). Microscopic observations were carried out on the yeast cells grown on cornmeal agar (CMA) and YPG agar.
To test for forcibly discharged ballistoconidia, these isolates were grown on cornmeal agar (CMA) (Merck, Germany). The dish was then inverted over another Petri dish of 5 % MEA and incubated at 28 °C. After 21 days of incubation in the inverted position, the MEA dish was checked for structures shot down from the colony grown on CMA. Microscopic observation was made from the above-mentioned cultures using Lactophenol Cotton Blue as a staining medium. Microscopic examinations were carried out using the 100 × objective of a Nikon Eclipse 80i microscope.
The physiological and biochemical characteristics of the isolates were determined according to the standard methods described by Barnett et al. (2000), while Kurtzman et al. (2011) were consulted to check whether the isolates represent already known species. To determine the enzymatic profile of the isolates, the degradation of starch, protein (casein), lipids (Tween 20 and Tween 80), phospholipids (egg yolk), gelatin, DNA, xylan, and carboxymethyl cellulose was studied according to the procedures from Brizzio et al. (2007). Petri plates containing each substrate were incubated at 28 °C.
To analyze the carbohydrate composition of the cell wall, 50 mg of dried cells cultivated from yeast extract peptone dextrose broth (YPD: 1 % yeast extract, 2 % peptone and 2 % dextrose (w/v)), were hydrolyzed with 1 ml of 1.0 M H2SO4 at 100 °C for 2 h, and then cooled, with the pH adjusted to 5.5 with a saturated solution of Ba(OH)2. The precipitate was removed by centrifugation. The supernatant was dried in a rotary evaporator and resolved in 50 μl of distilled water. A 1 μl aliquot of this solution was applied on a cellulose thin layer chromatography (TLC) plate and developed with a solvent system containing n-butanol: water: piridine: toluene (10:6:6:1) for 4 h. Sugars were visualized by an aniline phthalate solution and compared with a 1 μl aliquot of 1 % (w/v) of standard solutions of high purity sugar mixtures (Komagata and Suzuki 1987; Kurtzman et al. 2011).
DNA extraction, PCR, and sequencing
For DNA isolation, strains were grown in YPG broth at 28 °C for 24 h, followed by DNA extraction according to Hanna and Xiao (2006). ITS 1 and ITS 2 regions of the rDNA, including the 5.8S rDNA, were amplified by PCR using the primer pair ITS1 and ITS4 (White et al. 1990), the 5′-end of the nuclear large subunit ribosomal DNA (D1/D2 region, LSU) using the primer pair NL1 and NL4 (O’Donnell 1992, 1993), and the nuclear small subunit ribosomal DNA (SSU) using the primers NS1 and NS8 (White et al. 1990). DNA sequences determined for this study were deposited in GenBank, and accession numbers are given in Table 1.
Phylogenetic analyses
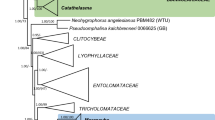
To elucidate the phylogenetic position of the isolated strains, their sequences were analysed within both single gene datasets (SSU, ITS, and LSU) and a concatenated dataset (SSU + LSU). Since preliminary analyses revealed an affinity of the two stains to the Urocystidales, the datasets were reduced to members of the Ustilaginomycetes, with some representatives of the Exobasidiomycetes as an outgroup. Each dataset covered all urocystidalean and ustilaginalean genera for which sequences were available in GenBank. If present in GenBank, the respective type species were used. Additionally, the sequence of an uncultured fungus clone, MAC09GadID (GenBank: JN890075), revealed by a Blast search (Altschul et al. 1997) as a closely related sequence, was added to the LSU dataset. GenBank accession numbers of the sequences used for the LSU dataset (Boekhout et al. 1995; Begerow et al. 1997, 2007; Piepenbring et al. 1999, 2002; Fell et al. 2000; Castlebury et al. 2005; Hendrichs et al. 2005; Stoll et al. 2005; Vánky et al. 2006, 2008a, 2013; Bauer et al. 2007, 2008; González et al. 2007; Matheny et al. 2007; McGuire et al. 2010; Vánky and Lutz 2011; Lutz et al. 2012a, b; McTaggart et al. 2012; Piątek et al. 2013) are given in Fig. 1.
Bayesian inference of phylogenetic relationships within the sampled Ustilaginomycetes: Markov chain Monte Carlo analysis of an alignment of LSU base sequences using the GTR+I+G model of DNA substitution with gamma distributed substitution rates and estimation of invariant sites, random starting trees and default starting parameters of the DNA substitution model. A 50 % majority-rule consensus tree is shown computed from 75,000 trees that were sampled after the process had become stationary. The topology was rooted with the exobasidiomycetous species Entyloma microsporum, Exobasidium vaccinii, and Tilletia caries. Support values greater than 70 are shown. Numbers on branches before slashes are estimates for a posteriori probabilities; numbers on branches after slashes are ML bootstrap support values. Branch lengths were averaged over the sampled trees. They are scaled in terms of expected numbers of nucleotide substitutions per site. The taxonomical concept applied corresponds to Begerow et al. (2014)
Sequence alignment was obtained using MAFFT 6.853 (Katoh et al. 2005; Katoh and Toh 2008) using the L-INS-i option. To obtain reproducible results, manipulations of the alignments by hand as well as manual exclusion of ambiguous sites were avoided, as suggested by Giribet and Wheeler (1999) and Gatesy et al. (1993), respectively. Instead, highly divergent portions of the alignments were omitted using GBlocks 0.91b (Castresana 2000), with the following options for the LSU data set. ‘Minimum Number of Sequences for a Conserved Position’: 28, ‘Minimum Number of Sequences for a Flank Position’: 28, ‘Maximum Number of Contiguous Non-conserved Positions’: 8, ‘Minimum Length of a Block’: 5, and ‘Allowed Gap Positions’ to ‘With half’.
The resulting alignments [LSU dataset: new number of positions: 602 (22 % of the original 2678 positions), number of variable sites: 333] were used for phylogenetic analyses using a Maximum Likelihood (ML) and a Bayesian Approach (BA) following Vasighzadeh et al. (2014).
For the LSU dataset, trees were rooted with the exobasidiomycetous species Entyloma microsporum (Unger) J. Schröt., Exobasidium vaccinii (Fuckel) Woronin, and Tilletia caries (DC.) Tul. & C. Tul.
Results
Morphological and physiological analyses
The growth rate and the colony appearance of the two isolates were different on the solid media tested (Table 2). One of the isolates (IBRC-M 30116T) produced cream-colored yeast colonies on YPG (Fig. 2a) and grey-colored yeast colonies on MEA, M40Y and DG18 (Fig. 2b–d), while the other isolate (IBRC-M 30112) produced cream-colored yeast colonies on all media (Fig. 2e–h). Both isolates produced colonies with eroded and fringed margins, except strain IBRC-M 30112, which produced rounded colonies on DG18. However, the two isolates were quite similar microscopically. The vegetative cells were mostly cylindrical, single or in small clusters. Vegetative reproduction via blastospores occurred by polar budding on short stalks (Fig. 3a). Ballistoconidia were produced on cornmeal agar (Fig. 3b) after 7 days of incubation at 28 °C. Ballistoconidia were formed terminally on sterigma-like structures in attachment to the hyphae or other ballistoconidia (Fig. 3c–d). Blastospore production was also observed in direct connection to the ballistoconidia in terminal or lateral position (Fig. 3e–f). Retraction septate hyphae and pseudohyphae were observed after 14 days of incubation on 5 % malt agar (Fig. 3g–i). Sexual structures and teliospore production were not observed in cultures of both strains. The detailed morphology is included in the species description and depicted in Figs. 2 and 3.
Micromorphology of Fereydounia khargensis (IBRC-M 30116T): a vegetative cells without or with blastospores produced by polar budding on short stalks (some budding cells indicated by arrows), b ballistoconidia on cornmeal agar, c–d formation of ballistoconidia (indicated by arrows) on sterigma-like structures in attachment to the hyphae or other ballistoconidium, e–f blastospore productions on ballistoconidia in terminal position and in lateral position, g retraction septate hypha, h–i pseudohyphae emerging from swollen vegetative cells on 5 % malt agar. Lactophenol Cotton Blue was used as a staining medium for c–f. Scale bar = 10 μm for a, b, g–i, and 20 μm for c–f
Fermentation ability was absent. Assimilation of most of carbon sources was observed. Based on the enzymatic activity assay, the isolates produced urease, gelatinase, and lipase. DNase production was observed for only one of the isolates (IBRC-M 30112). The TLC chromatogram from the whole cell hydrolysate of sugars indicated the predominance of glucose and the absence of xylose.
Phylogenetic analyses
The ITS sequences of the two isolates were identical, LSU sequences differed in 2 bp (0.34 %), and SSU sequences differed in 8 bp (0.47 %). Analyses of all datasets revealed the two isolates within the Urocystidales. The different runs of the BA that were performed and the ML analyses yielded consistent topologies. To illustrate the results, the consensus tree of one run of the BA of the LSU dataset is presented (Fig. 1), because the LSU dataset covered substantially more genera compared to both the ITS and SSU datasets.
Using the exobasidiomycetous species as outgroup, the statistically well-supported groups appearing in the phylogenetic tree were congruent to the families and orders discussed in Begerow et al. (2014). In all analyses, the sequences of the two isolates clustered together within the Urocystidales, forming a lineage together with Mycosyrinx cissi (DC.) Beck (Mycosyringaceae), an uncultured fungus clone MAC09GadID (GenBank: JN890075), and the clade including the Floromycetaceae and Urocystidaceae. However, the relation between those four groups received only low support values.
Taxonomy
Fereydouniaceae S. Nasr, M.R. Soudi, H.D.T. Nguyen, M. Lutz & M. Piątek, fam. nov.
MycoBank No. MB 809914
Description: Members of the order Urocystidales according to analyses of SSU, ITS and LSU loci, developing a saprobic anamorphic stage that produces ballistoconidia, and lacking sexual structures and teliospores.
Type genus: Fereydounia S. Nasr, M.R. Soudi, H.D.T. Nguyen, M. Lutz, M. Piątek 2014.
Fereydounia S. Nasr, M.R. Soudi, H.D.T. Nguyen, M. Lutz & M. Piątek, gen. nov.
MycoBank No. MB 809915
Etymology: Fereydounia, named in the honor of the late Prof. Fereydoun Malekzadeh (1933–2012) for his contribution to microbiology in Iran.
Description: Members of the Fereydouniaceae with a predominance of glucose and an absence of xylose in whole-cell hydrolysates, a broad spectrum of carbon source assimilation, urease activity, absence of fermentation ability, and a positive diazonium blue B reaction. Ballistoconidia, blastospores, retraction septate hyphae and pseudohyphae are present. Sexual structures and teliospore production absent.
Type species: Fereydounia khargensis S. Nasr, M.R. Soudi, H.D.T. Nguyen, M. Lutz & M. Piątek 2014.
Fereydounia khargensis S. Nasr, M.R. Soudi, H.D.T. Nguyen, M. Lutz & M. Piątek, sp. nov. Figs. 2 and 3
MycoBank No. MB 809916
Etymology: khargensis, in reference to the Kharg Island where the species was found.
Type: Iran, Bushehr Province, Kharg Island, from undetermined plant remnants, 1 August 2013, leg. S. Nasr & M.R. Soudi [holotype: IBRC-M 30116T preserved in a metabolically inactive state (lyophilized); CBS 13305T – Isotype].
Description: Colony diameters (mm) after 7 days at 28 °C: MEA 16–21 (mean = 14.5, n = 9); YPG 18–22 (mean = 20, n = 9); M40Y 8–11 (mean = 13.0, n = 9); DG18 20–30 (mean = 25, n = 9). The isolate produced cream-colored yeast colonies on YPG (Fig. 2a) and grey-colored yeast colonies on MEA (Fig. 2b), M40Y (Fig. 2c) and DG18 (Fig. 2d) with eroded and fringed margins. Vegetative reproduction via blastospores occurred by polar budding on short stalks (Fig. 3a). Production of ballistoconidia observed on cornmeal agar. On cornmeal agar, ballistoconidia were produced, 5.5–15.5 × 1.5–3.5 μm (mean ± SD = 10.0 ± 0.7 × 2.0 ± 0.1 μm), single (Fig. 3b) or in small clusters. Ballistoconidia were formed terminally on sterigma-like structures in attachment to the hyphae or other ballistoconidia (Fig. 3c–d). Blastospore production was also observed in direct connection to the ballistoconidia in terminal or lateral position (Fig. 3e–f). The retraction septate hyphae and pseudohyphae emerging from swollen vegetative cells (Fig. 3g–i) were observed after 14 days of incubation on 5 % malt agar. Sexual structures were not observed. Fermentation ability was absent. The following carbon compounds were assimilated: D-glucose, D-galactose, sucrose, maltose, cellobiose, trehalose, raffinose, lactose, melibiose, melezitose, D-xylose, L-arabinose, D-arabinose, D-ribose, L-rhamnose, soluble starch, ethanol, glycerol, ribitol, D-mannitol, myo-Inositol, salicin (weak), succinic acid, citric acid, DL-lactic acid, n-hexadecane, arbutin; no growth occured on inulin and methanol. The nitrogen compounds potassium nitrate, sodium nitrite, lysine, ethylamine, and cadaverine were assimilated and imidazole, creatine and creatinine were not assimilated. Growth occurred at 34 and 37 °C but not at 40 °C. Growth occurred in vitamin-free medium, and media containing 50 and 60 % glucose. Growth occurred in the presence of 0.01 % cycloheximide, but not in 0.1 % cycloheximide. Growth occurred in the presence of 10 % sodium chloride, but not in 16 % sodium chloride. No starch-like substances were produced. Diazonium blue B reaction and urease activity were positive.
Discussion
The morphological, physiological and molecular phylogenetic analyses conducted in this study revealed that the two strains of yeast-like fungus isolated from plant remains on Kharg Island in Iran belong to an undescribed genus and species that is named here as Fereydounia khargensis. Molecular phylogenetic reconstructions inferred that, within the Ustilaginomycotina, this species resides in the Urocystidales, a distinct order currently including teliosporic species (classical smut fungi) parasitic on dicotyledonous and monocotyledonous host plants (Vánky 2012). In recent classification schemes (Begerow et al. 2014), the order Urocystidales is split into five main lineages assigned to families: the Glomosporiaceae, containing the single genus Thecaphora with 63 species parasitic on dicotyledonous plants from 16 host families (Vánky et al. 2008b; Vánky 2012; Vasighzadeh et al. 2014); the Doassansiopsidaceae, including the single genus Doassansiopsis (Setch.) Dietel with 14 species parasitic on dicot and monocot plants from five host families (Piątek et al. 2008; Vánky 2012); the Mycosyringacae, containing the single genus Mycosyrinx Beck with four species parasitic on Vitaceae (Vánky 2012); the Floromycetaceae, accommodating two genera, Floromyces Vánky, M. Lutz & R. Bauer with one species and Antherospora R. Bauer, M. Lutz, Begerow, M. Piątek & Vánky with 11 species, both parasitic on Asparagaceae (Vánky et al. 2008a; Piątek et al. 2011, 2013; Vánky 2012); and the Urocystidaceae, including seven genera: Flamingomyces R. Bauer, M. Lutz, M. Piątek, Vánky & Oberw. with one species on Ruppiaceae, Melanoxa M. Lutz, Vánky & R. Bauer with two species on Oxalidaceae, Melanustilospora Denchev with two species on Araceae, Mundkurella Thirum. with five species on Araliaceae, Urocystis Rabenh. ex Fuckel with 166 species on different monocot and dicot families, Ustacystis Zundel with one species on Rosaceae, and Vankya Ershad with three species on Liliaceae (Denchev 2003; Bauer et al. 2007; Denchev and Kakishima 2007; Vánky 2009a, 2009b, 2012; Vánky and Abbasi 2011; Lutz et al. 2012a).
Molecular phylogenetic analyses conducted in this study revealed that Fereydounia khargensis represents a distinct urocystidalean lineage that cannot be assigned to any known family. Therefore, the new family Fereydouniaceae is introduced to accommodate Fereydounia khargensis. Morphologically, Fereydouniaceae is unique within Urocystidales, in that it accommodates an anamorphic species living freely in the environment and apparently not connected to any living host plant. It is currently unclear whether Fereydounia khargensis is a species that lost the ability to produce a dikaryotic stage or if the dikaryotic phase is simply undetected yet. However, considering that plant parasitic teliosporic species are relatively well known in southern Asia (and therefore a unique teleomorphic urocystidalean genus should already have been detected), and also that anamorph–teleomorph connections are in fact only rarely detected in molecular studies on the ustilaginomycotinous fungi [e.g., probable pairs may be Farysizyma acheniorum (Buhagiar & J.A. Barnett) Á. Fonseca–Farysia thuemenii (A.A. Fisch. Waldh.) Nannf., Inácio et al. 2008; Pseudozyma prolifica–Ustilago maydis (DC.) Corda, Pseudozyma tsukubaensis (Onishi) Boekhout–Macalpinomyces spermophorus (Berk. & M.A. Curtis ex de Toni) Vánky, Begerow et al. 2000; Sampaio 2004; Rhodotorula bacarum (Buhagiar) Rodr. Mir. & Weijman–Microstroma album (Desm.) Sacc., de Beer et al. 2006], it is tempting to speculate that Fereydounia khargensis represents an anamorphic saprobic species that lost the ability to produce a dikaryotic phase.
Although Fereydounia khargensis is the first saprobic anamorph in the Urocystidales, asexual stages were already reported in some teliosporic plant parasitic members of this order. In contrast to Fereydounia khargensis, they were, however, always observed on the same living host plant as the teleomorph, and apparently, development of the anamorphic parasitic stage precedes development of the teliosporic stage; for example, in Thecaphora lathyri J.G. Kühn, T. seminis-convolvuli (Desm.) Ito, T. thlaspeos (Beck) Vánky, Urocystis primulae (Rostr.) Vánky or U. trientalis (Berk. & Broome) B. Lindeb. (Vánky 1994). An anamorphic parasitic stage was also observed in Thecaphora saponariae (F. Rudolphi) Vánky s.l. on Silene colorata Poir., though the teliosporic stage has not been subsequently observed on this host (Vánky and Lutz 2007).
Fereydounia khargensis may represent a part of a larger diversity of urocystidalean saprobic yeast-like fungi. In the molecular phylogenetic analyses, the uncultured fungus clone MAC09GadID from an environmental soil sample in mixed lowland tropical rainforest in Guyana (GenBank: JN890075, generated for McGuire et al. 2010 but not included in a publication) clusters within Urocystidales, forming another distinct lineage. However, the formal description of this fungus needs isolation and culture of an appropriate organism that could be described and illustrated morphologically.
The discovery of a saprobic yeast fungus representing a new lineage in the order Urocystidales contributes greatly to the knowledge on the evolutionary diversity in the Ustilaginomycotina. With this finding, the number of ustilaginomycotinous orders with still unknown free-living anamorphic species, is reduced to three—Ceraceosorales, Doassansiales, and Tilletiales. However, with the increasing application of molecular methods to identify yeast isolates, this may change, and novel lineages could be discovered, especially amongst the strains isolated from still poorly explored tropical and subtropical ecosystems.
References
Afsah-Hejri L (2013) Saprophytic yeasts: effective biocontrol agents against Aspergillus flavus. Int Food Res J 20:3403–3409
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Barnett JA, Payne RW, Yarrow D (2000) Yeasts: characteristics and identification, 3rd edn. Cambridge University Press, Cambridge
Bauer R, Oberwinkler F, Vánky K (1997) Ultrastructural markers and systematics in smut fungi and allied taxa. Can J Bot 75:1273–1314
Bauer R, Begerow D, Nagler A, Oberwinkler F (2001a) The Georgefischeriales: a phylogenetic hypothesis. Mycol Res 105:416–424
Bauer R, Begerow D, Oberwinkler F, Piepenbring M, Berbee ML (2001b) Ustilaginomycetes. In: McLaughlin DJ, McLaughlin EG, Lemke PA (eds) The Mycota. Vol. VII. Part B. Systematics and evolution. Springer, Berlin, pp 57–83
Bauer R, Lutz M, Oberwinkler F (2005) Gjaerumia, a new genus in the Georgefischeriales (Ustilaginomycetes). Mycol Res 109:1250–1258
Bauer R, Begerow D, Sampaio JP, Weiß M, Oberwinkler F (2006) The simple-septate basidiomycetes: a synopsis. Mycol Prog 5:41–66
Bauer R, Lutz M, Piątek M, Vánky K, Oberwinkler F (2007) Flamingomyces and Parvulago, new genera of marine smut fungi (Ustilaginomycotina). Mycol Res 111:1199–1206
Bauer R, Lutz M, Begerow D, Piątek M, Vánky K, Bacigálová K, Oberwinkler F (2008) Anther smut fungi on monocots. Mycol Res 112:1297–1306
Begerow D, Bauer R, Oberwinkler F (1997) Phylogenetic studies on nuclear LSU rDNA sequences of smut fungi and related taxa. Can J Bot 75:2045–2056
Begerow D, Bauer R, Boekhout T (2000) Phylogenetic placements of ustilaginomycetous anamorphs as deduced from nuclear LSU rDNA sequences. Mycol Res 104:53–60
Begerow D, Stoll M, Bauer R (2007 (2006)) A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia 98:906–916
Begerow D, Schäfer AM, Kellner R, Yurkov A, Kemler M, Oberwinkler F, Bauer R (2014) Ustilaginomycotina. In: McLaughlin DJ, Spatafora JW (eds) The Mycota. vol. 7. Part A. Systematics and evolution, 2nd edn. Springer, Berlin
Boekhout T, Fell JW, O’Donnell K (1995) Molecular systematics of some yeast-like anamorphs belonging to the Ustilaginales and Tilletiales. Stud Mycol 38:175–183
Boekhout T, Theelen B, Houbraken J, Robert V, Scorzetti G, Gafni A, Gerson U, Sztejnberg A (2003) Novel anamorphic mite-associated fungi belonging to the Ustilaginomycetes: Meira geulakonigii gen. nov., sp. nov., Meira argovae sp. nov. and Acaromyces ingoldii gen. nov., sp. nov. Int J Syst Evol Microbiol 53:1655–1664
Boekhout T, Gildemacher P, Theelen B, Müller WH, Heijne B, Lutz M (2006) Extensive colonization of apples by smut anamorphs causes a new postharvest disorder. FEMS Yeast Res 6:63–76
Brizzio S, Turchetti B, de García V, Libkind D, Buzzini P, van Broock M (2007) Extracellular enzymatic activities of basidiomycetous yeasts isolated from glacial and subglacial waters of northwest Patagonia (Argentina). Can J Microbiol 53:519–525
Castlebury LA, Carris LM, Vánky K (2005) Phylogenetic analysis of Tilletia and allied genera in order Tilletiales (Ustilaginomycetes; Exobasidiomycetidae) based on large subunit nuclear rDNA sequences. Mycologia 97:888–900
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
de Beer ZW, Begerow D, Bauer R, Pegg GS, Crous PW, Wingfield MJ (2006) Phylogeny of the Quambalariaceae fam. nov., including important Eucalyptus pathogens in South Africa and Australia. Stud Mycol 55:289–298
Denchev CM (2003) Melanustilospora, a new genus in the Urocystales (smut fungi). Mycotaxon 87:475–477
Denchev CM, Kakishima M (2007) Notes on some Japanese smut fungi. IV. Mundkurella japonica, sp. nov. Mycotaxon 102:9–16
Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A (2000) Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol 50:1351–1371
Gatesy J, DeSalle R, Wheeler W (1993) Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Mol Phylogenet Evol 2:152–157
Giribet G, Wheeler WC (1999) On gaps. Mol Phylogenet Evol 13:132–143
González V, Vánky K, Platas G, Lutz M (2007) Portalia gen. nov. (Ustilaginomycotina). Fungal Divers 27:45–59
Hanna M, Xiao W (2006) Isolation of nucleic acids. In: Xiao W (ed) Yeast Protocols (Methods in Molecular Biology), Volume 313. Humana Press, pp 15–20
Hedayati MT, Hajheydari Z, Hajjar F, Ehsani A, Shokohi T, Mohammadpour R (2010) Identification of Malassezia species isolated from Iranian seborrhoeic dermatitis patients. Eur Rev Med Pharmacol Sci 14:63–68
Hendrichs M, Begerow D, Bauer R, Oberwinkler F (2005) The genus Anthracoidea (Basidiomycota, Ustilaginales): a molecular phylogenetic approach using LSU rDNA sequences. Mycol Res 109:31–40
Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Lumbsch HT, Lutzoni F, Matheny PB, Mclaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miądlikowska J, Miller A, Moncalvo JM, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schüßler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiß M, White MM, Winka K, Yao YJ, Zhang N (2007) A higher-level phylogenetic classification of the Fungi. Mycol Res 111:509–547
Inácio J, Landell MF, Valente P, Wang PH, Wang YT, Yang SH, Manson JS, Lachance MA, Rosa CA, Fonseca Á (2008) Farysizyma gen. nov., an anamorphic genus in the Ustilaginales to accommodate three novel epiphytic basidiomycetous yeast species from America, Europe and Asia. FEMS Yeast Res 8:499–508
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program (outlines version 6). Brief Bioinform 9:286–298
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Komagata K, Suzuki KI (1987) Lipid and cell wall analysis in bacterial systematic. In: Colwell RR, Grigorova & (eds) Methods in Microbiology. Volume 19. Current Methods for Classification and Identification of Microorganisms. Academic Press, pp 201–203
Kurtzman CP, Fell JW, Boekhout T (2011) The yeasts, a taxonomic study, 5th edn. Elsevier Science Publication, Amsterdam
Lutz M, Vánky K, Bauer R (2012a) Melanoxa, a new genus in the Urocystidales (Ustilaginomycotina). Mycol Prog 11:149–158
Lutz M, Vánky K, Piątek M (2012b) Shivasia gen. nov. for the Australasian smut Ustilago solida that historically shifted through five different genera. IMA Fungus 3:143–154
Matheny PB, Gossmann JA, Zalar P, Arun Kumar TK, Hibbett DS (2007 (2006)) Resolving the phylogenetic position of the Wallemiomycetes: an enigmatic major lineage of Basidiomycota. Can J Bot 84:1794–1805
McGuire KL, Zak DR, Edwards IP, Blackwood CB, Upchurch R (2010) Slowed decomposition is biotically mediated in an ectomycorrhizal, tropical rain forest. Oecologia 164:785–795
McTaggart AR, Shivas RG, Geering ADW, Callaghan B, Vánky K, Scharaschkin T (2012) Soral synapomorphies are significant for the systematics of the Ustilago-Sporisorium-Macalpinomyces complex (Ustilaginaceae). Persoonia 29:63–77
Moniri R, Nazeri M, Amiri S, Asghari B (2009) Isolation and identification of Malassezia spp. in pytiriasis versicolor in Kashan, Iran. Pak J Med Sci 25:837–840
Nguyen HDT, Nickerson NL, Seifert KA (2013) Basidioascus and Geminibasidium: a new lineage of heat resistant and xerotolerant basidiomycetes. Mycologia 105:1231–1250
O’Donnell KL (1992) Ribosomal DNA internal transcribed spacers are highly divergent in the phytopathogenic ascomycete Fusarium sambucinum (Gibberella pulicaris). Curr Genet 22:213–220
O’Donnell KL (1993) Fusarium and its near relatives. In: Reynolds DR, Taylor JW (eds) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, pp 225–233
Piątek M, Vánky K, Mossebo DC, Piątek J (2008) Doassansiopsis caldesiae sp. nov. and Doassansiopsis tomasii: two remarkable smut fungi from Cameroon. Mycologia 100:662–672
Piątek M, Lutz M, Smith PA, Chater AO (2011) A new species of Antherospora supports the systematic placement of its host plant. IMA Fungus 2:135–142
Piątek M, Lutz M, Chater AO (2013) Cryptic diversity in the Antherospora vaillantii complex on Muscari species. IMA Fungus 4:5–19
Piepenbring M, Begerow D, Oberwinkler F (1999) Molecular sequence data assess the value of morphological characteristics for phylogenetic classification of Cintractia. Mycologia 91:485–498
Piepenbring M, Stoll M, Oberwinkler F (2002) The generic position of Ustilago maydis, Ustilago scitaminea, and Ustilago esculenta (Ustilaginales). Mycol Prog 1:71–80
Sampaio JP (2004) Diversity, phylogeny and classification of basidiomycetous yeasts. In: Agerer R, Blanz P, Piepenbring M (eds) Frontiers in basidiomycote mycology. IHW-Verlag, Eching, pp 49–80
Shokohi T, Afshar P, Barzgar A (2009) Distribution of Malassezia species in patients with pityriasis versicolor in Northern Iran. Indian J Med Microbiol 27:321–324
Simpson JA (2000) Quambalaria, a new genus of eucalypt pathogens. Australas Mycol 19(2):57–62
Sipiczki M, Kajdacsi E (2009) Jaminaea angkorensis gen. nov., sp. nov., a novel anamorphic fungus containing an S943 nuclear small-subunit rRNA group IB intron represents a basal branch of Microstromatales. Int J Syst Evol Microbiol 59:914–920
Stoll M, Begerow D, Oberwinkler F (2005) Molecular phylogeny of Ustilago, Sporisorium, and related taxa based on combined analyses of rDNA sequences. Mycol Res 109:342–356
Sugiyama J, Tokuoka K, Suh SO, Hirata A, Komagata K (1991) Sympodiomycopsis: a new yeast-like anamorph genus with basidiomycetous nature from orchid nectar. Antonie Van Leeuwenhoek 59:95–108
Vánky K (1994) European smut fungi. G. Fischer Verlag, Stuttgart
Vánky K (2009a) Ustacystis waldsteiniae, a remarkable smut fungus (Ustilaginomycetes). Mycol Balc 6:67–72
Vánky K (2009b) The genus Vankya (Urocystidaceae) revisited. Mycol Balc 6:73–78
Vánky K (2012) Smut fungi of the world. APS Press, St. Paul
Vánky K, Abbasi M (2011) Urocystis phalaridis sp. nov. (Urocystidaceae) on Phalaris sp. (Poaceae) from Iran. Rostaniha 12:187–190
Vánky K, Abbasi M (2013) Smut fungi of Iran. Mycosphere 4(3):363–454
Vánky K, Lutz M (2007) Revision of some Thecaphora species (Ustilaginomycotina) on Caryophyllaceae. Mycol Res 111:1207–1219
Vánky K, Lutz M (2011) Tubisorus, a new genus of smut fungi (Ustilaginomycetes) for Sorosporium pachycarpum. Mycol Balc 8:129–135
Vánky K, Lutz M, Shivas RG (2006) Anomalomyces panici, new genus and species of Ustilaginomycetes from Australia. Mycol Balc 3:119–126
Vanky K, Lutz M, Bauer R (2008a) Floromyces, a new genus of Ustilaginomycotina. Mycotaxon 104:171–184
Vánky K, Lutz M, Bauer R (2008b) About the genus Thecaphora (Glomosporiaceae) and its new synonyms. Mycol Prog 7:31–39
Vánky K, Shivas RG, Barrett MD, Lutz M (2013) Eriocortex eriocauli, gen. et sp. nov. (Ustilaginomycetes) from Australia. Mycobiota 1:9–16
Vasighzadeh A, Zafari D, Selçuk F, Hüseyin E, Kurşat M, Lutz M, Piątek M (2014) Discovery of Thecaphora schwarzmaniana on Rheum ribes in Iran and Turkey: implications for the diversity and phylogeny of leaf smuts on rhubarbs. Mycol Prog 13:881–892
Wang QM, Theelen B, Groenewald M, Bai FY, Boekhout T (2014) Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Persoonia 33:41–47
White TJ, Bruns TD, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR Protocols: a guide to methods and applications. Academic, San Diego, pp 315–322
Zarei-Mahmoudabadi A, Zarrin M, Mehdinezhad F (2013) Seborrheic dermatitis due to Malassezia species in Ahvaz, Iran. Iran J Microbiol 5:268–271
Acknowledgments
This research was financially supported by the Iranian Biological Resource Center (IBRC), ACECR and we owe a great amount of thanks to Mohammad Ali Amoozegar (Head of Department) for his support.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Nasr, S., Soudi, M.R., Shahzadeh Fazeli, S.A. et al. Expanding evolutionary diversity in the Ustilaginomycotina: Fereydouniaceae fam. nov. and Fereydounia gen. nov., the first urocystidalean yeast lineage. Mycol Progress 13, 1012 (2014). https://doi.org/10.1007/s11557-014-1012-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-014-1012-0