Abstract
Purpose
As the most common primary intracranial tumor, glioblastoma (GBM) is a malignant tumor that originated from neuroepithelial tissue, accounting for 40–50% of brain tumors. Precise survival prediction for patients suffering from GBM can not only help patients and doctors formulate treatment plans, but also help researchers understand the development of the disease and stimulate medical development.
Methods
In view of the tedious process of manual feature extraction and selection in traditional radiomics, we propose an end-to-end survival prediction model based on DenseNet to extract the features of magnetic resonance images including T1-weighted post-contrast images and T2-weighted images through two-branch networks. After segmenting the region of interest, the original image, the image of tumor region and the image without tumor are combined as input sample sets with three channels. Additionally, for some patients having only one of T1- or T2-weighted images, One2One CycleGAN is used to generate the T1 image from the T2 image, or vice versa. Flipping and rotating are also used for sample augmentation.
Result
By using the augmented training sample set to train the model, the classification and prediction accuracy of the two-branch DenseNet survival prediction model can reach up to 94%, and the Kaplan–Meier survival curve indicates that the model can classify patients into high-risk group and low-risk group based on whether they could survive for more than three years.
Conclusion
The classification and prediction results of the model and the survival analysis demonstrate that our model can get superior classification results which can be referenced by doctors and patients’ families for developing medical plans. However, improving the loss function and expanding the sample size can further improve the prediction results, which are the target of our subsequent research.
Similar content being viewed by others
Introduction
Glioblastoma (GBM) is one of the most common brain tumors, which has the characteristics of high morbidity, high recurrence, high mortality and low cure rate [1]. Therefore, a prediction of the survival time of glioma patients can be a reference for the formulation of diagnosis and treatment plans, which can improve the diagnosis efficiency and save medical resources. As we known, three-year survival refers to the proportion of a tumor that survives more than three years after comprehensive treatment and is usually used to indicate the efficacy of various cancers [2]. According to the 3-year survival time of patients suffering from GBM in the TCIA database [3], the overall and 3-year survival rates are shown in Table 1.
The statistical analysis of the samples in the TCIA database shows that the probability of survival of a GBM patient is 79.7% when the survival period of GBM patients is more than three years. It indicates that patients with a survival period of more than three years are more likely to survive, that is, these patients belong to the low-risk group, and the others belong to the high-risk group.
As we know, the diagnosis of tumor mainly depends on the analysis of medical images, such as magnetic resonance images (MRI), including T1-weighted images and T2-weighted images. Therefore, when making survival prediction on tumor patients, the common procedure is to manually extract and filter medical images’ features before using the classification method of machine learning to predict the survival time of the patients. Chang et al. [4] generated the model with volumetric, shape, texture, parametric and histogram features derived from multimodal MRI and used random forest classifier in machine learning method to predict survival in recurrent GBM patients treated with bevacizumab. Osman [5] extracted patient’s age, segmented tumor’s size and its location features; and used support vector machines in machine learning method to develop the algorithm for predicting the overall survival for GBM patients. The algorithm was trained, validated and tested on BRATS’2017’s training, validation, and testing data sets. Meanwhile, for other tumors such as non-small cell lung cancer, the prediction of survival outcomes could also be investigated based on radiomics texture and shape features extracted from CT scans. And, automatic random forest classification was used to predict patient survival groups from multivariate feature data [6]. In recent years, deep learning methods have provided state-of-the-art performance in the field of medical image processing, such as image segmentation [7], classification [8] and survival prediction [9]. Research on survival prediction based on deep learning methods continues to emerge. A dense motion model using a fully convolutional network for survival prediction of patients diagnosed with pulmonary hypertension was created by Bello et al. [10] using image sequences of the heart, acquired via cardiac MRI. This work demonstrated how a complex computer vision task using high-dimensional medical image data can efficiently predict human survival. Another deep learning model that combined the information from three different kinds of sources, which are clinical characteristics, MRI and brain morphology data, increased the amyotrophic lateral sclerosis patients’ survival prediction accuracy to 84.4% [11]. Nie et al. [12] proposed a two-stage learning-based method to predict the overall survival time of high-grade gliomas patients. The experimental results from their study demonstrated that this multi-model, multi-channel deep survival prediction framework achieved an accuracy of 90.66%, outperforming the competing machine learning methods such as support vector machine. A deep learning framework was also used by Tabibu et al. for the classification of renal cell carcinoma subtypes, and for identification of features that predicted survival outcome from digital histopathological images [13].
Similar to the aforementioned research on the survival prediction of tumor patients using deep learning method, based on DenseNet [14], which is an effective classification network in deep learning, we propose a novel two-branch survival prediction model. In this model, post-contrast T1-weighted images and corresponding T2-weighted images are preprocessed into three-channel sample sets and respectively sent into the two branches. Here, the three-channel samples are combined by the whole brain image, the image only with tumor area, and the image without tumor. Furthermore, in order to reduce overfitting, a virtual sample generation method based on One2One CycleGAN [15] is also used to complement the insufficient medical images besides traditional augmentation method including flipping and rotation.
Methods
Virtual samples generating
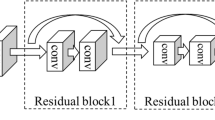
Generation adversarial network (GAN) [16] is usually used for generating virtual samples in deep learning. Due to the lack of T1- or T2-weighted MRI images of some patients in the database, One2One CycleGAN which is an improved novel algorithm in GAN is used to generate T2-weighted images from corresponding post-contrast T1-weighted images, or vice versa. Different from CycleGAN [17] or pix2pix [18] using two one-way generators for the translation between post-contrast T1 images and T2 images, only one generator based on residual network (ResNet) [19] is used for both tasks for bidirectional unpaired or paired image-to-image translation as shown in Fig. 1.
The One2One CycleGAN method has two separate losses, one for each image transfer direction, and L1-norm is used instead of L2-norm for less blurring of the image. For the mapping function G: T1 → T2 and its discriminator DY, the adversarial loss, cycle consistency loss and the final objective are as follows [15]:
Similarly, the mapping function G: T2 → T1 and its discriminator DX, the adversarial loss, cycle consistency loss and the final objective are:
By solving the minimax optimization problems, we can get the satisfactory generator (G) and discriminators (DY and DX).
In order to reduce overfitting caused by insufficient samples, besides generating virtual samples, traditional augmentation methods like flipping and rotating implemented by Augmentor [20] are also used to increase the number of training samples. Subsequent experiments will compare the classification results with and without virtual samples.
Two-branch DenseNet survival prediction model
To automatically extract image features derived from deep learning for survival prediction, based on DenseNet, we propose a novel architecture of a two-branch network as shown in Fig. 2. In this network, each branch learns the features of post-contrast T1 and T2 images separately through DenseNet, and the combination of the final layers of each branch is sent to the classifier for classification and prediction. Specifically, the input samples are firstly operated by 3 × 3 convolution layers, and then image features are extracted by three dense blocks and three transition layers. After that, a classification result with a size of 2 × (batch size) can be obtained.
We designed software to manually crop the region of interest of an image. For each T1 post-contrast image or T2 image used in this study, three types of images with the same size of 256 × 256 are obtained: the original image, the tumor image and the image without tumor, which are combined into one 3-channel sample as shown in Fig. 2a. The advantage of this method is to retain the location and the size of the tumor in the brain. Then, in every iteration, samples with a size of 256 × 256 × 3 × (batch size) are sent to each branch of the model for training.
DenseNet usually consists of several dense blocks and transition layers. The dense block defines the connection between input and output while the transition layer controls the number of channels. The transition layer is represented as a trapezoid in Fig. 2b, which is usually composed of batch normalization (BN), followed by a Rectified Linear Unit (ReLu), 2 × 2 pooling and 1 × 1 convolution operations. The dense block is the most characteristic part of the DenseNet, and its architecture is shown in Fig. 3.
In one dense block, all previous layers are connected as an input to the next layer:
Hi in formula (7) represents a nonlinear transformation, which is a combined operation including BN, ReLu and 3 × 3 convolution operations. Each layer receives feature maps from all of the previous layers, so the network can be thinner and more compact. While the classifier uses features from all levels, it tends to give smoother decision boundaries, making DenseNet perform better when the training data is insufficient. Besides, error signals could be propagated more directly into earlier layers. This is an implicit deep supervision, because early layers can get supervision directly from the final classification layer.
Last but not least, in this end-to-end survival prediction model, the feature extraction and classification are integrated into a whole, and the softmax loss functions are added both at the end of each branch and the end of the model, which ensures that every branch can be fully trained to avoid the local optimal solution of the model.
Experiment and result
Virtual samples generating
Due to the lack of one of T1 and T2 images for some patients in TCIA database, One2One CycleGAN method is used to generate the virtual T1 or T2 images for the following study. To prove the feasibility of this method, two experiments are conducted with BraTS 2018 data set [21,22,23] and TCIA data set separately.
Data
The BraTS 2018 data set offers medical images of 210 glioblastoma patients for training and 68 for testing. Four types of MR images of each patient were collected: native (T1), post-contrast T1-weighted (T1Gd), T2-weighted (T2), and T2 Fluid Attenuated Inversion Recovery (FLAIR). Each type of MR image’s 3D volumetric scan contains 155 2D slices. T1Gd and T2 images were chosen to make the sample sets. 2D slices with tumor features were selected from the chosen MR 3D scan images of each patient, and a total of 13,203 2D slice pairs (T1Gd and T2 images) were achieved for training, 4293 for testing. The images are resized to 128 × 128. In TCIA database, after manual screening, the slices with tumor characteristics are retained. The number of slices kept for each patient is not fixed. A total of 356 sample pairs with a size of 256 × 256 are selected, 310 for training and 46 for testing.
Result
In the experiment conducted with BraTS 2018 data set, the loss weights are set as λx = λy = 10, the batch size is set to 10, and the learning rate is fixed for the first 100 epochs and linearly decayed to zero over the next 100 epochs. Figure 4 shows some examples of virtual T1 and T2 images generated by one2one CycleGAN.
Similarly, in TCIA database, the loss weights are set as λx = λy = 10, the batch size is set to 2. Some original images and generated virtual images are shown in Fig. 5. In this paper, TCIA-GBM refers to the TCGA-GBM medical imaging collection in the TCIA database corresponding to the cases in the TCGA gene database. In order to avoid confusion with the gene set, it is abbreviated as TCIA-GBM.
Comparing the results using different data sets, the PSNR and SSIM [24] of the generated images are shown in Table 2. Higher values of PSNR and SSIM indicate better quality of the generated virtual images.
Experimental results in Figs. 4, 5 and Table 2, indicate that the generated virtual images well retain most of the characteristics of the original images, especially the outline of the original image, the gray values of different tissues and the location of the tumor. In particular, the virtual generation of T2-weighted images being more successful than that of T1-weighted images regarding the tumor features. It is likely that the tumor tissue has high contrast and prominent features relative to the background in the post-contrast T2 images. Meanwhile, it is also noticed that PSNR and SSIM of BraTS data set outperform that of TCIA data set, which could be caused by the sample size. In the following work, it is necessary to collect more samples of patients suffering from GBM.
Survival prediction and classification
Data
In the experiment of survival prediction and classification, in order to keep the sample size consistent between the groups of patients surviving less than three years and more than three years, only 250 MRI image pairs of 60 patients suffering from GBM (out of 356 image pairs mentioned in “Virtual samples generating” section) were selected from the TCIA database, including T1-weighted post-contrast images and T2-weighted images. Two-thirds of the patients were male and ranged in age from 14 to 86, all patient information is anonymous. Half of these patients have a survival period of more than three years and the others of less than three years. Fivefold cross-validation was used to comprehensively and quantitatively evaluate the two-branch DenseNet survival prediction model. The sample set is randomly partitioned into five equal-sized and non-overlap subsets, the image slices of the same patient will not appear in different subsets. Of the five subsets, a single subset is retained as the testing data for testing the model, and the remaining four subsets (three subsets as training set and one as validation set) are used as training data. The cross-validation process is then repeated five times, with each of the five subsets used exactly once as the testing data.
After virtual samples generated by One2One CycleGAN and samples augmented by flipping and rotating through Augmentor, in order to evaluate the influence of the number and type of samples on the experimental results, four sample sets were used in the experiments:
-
1.
Sample I: 250 original sample pairs (T1 and corresponding T2 samples);
-
2.
Sample II: 250 original sample pairs and 1000 augmented samples generated by flipping and rotating the samples in training set, totally 1250 sample pairs;
-
3.
Sample III: 250 original sample pairs, 92 paired samples generated by One2One CycleGAN from 46 original T1 images and 46 original T2 images as supplement of training set, totally 342 sample pairs;
-
4.
Sample IV: 342 sample pairs from Sample III and 1000 augmented samples generated by flipping and rotating the samples in training set, totally 1342 sample pairs.
Result
In the two-branch DenseNet survival prediction experiment, the training is implemented on a personal computer equipped with an Intel i5-8400 CPU and a NVIDIA 1080 GPU. Adam optimizer is used for training with a batch size of 8, the training epoch is set to 50, and an initial learning rate of 0.001 linearly decayed to zero.
There are several objective criteria [25,26,27] for the two-category problem to evaluate the prediction quality of the model: (1) Accuracy, (2) Recall, (3) Precision, (4) F1 score, (5) ROC curve (receiver operator characteristic curve) and (6) AUC (area under curve). Table 3 shows the classification results of different sample sets used in the training of two-branch DenseNet survival prediction model. Higher values of accuracy, recall, precision and F1 score indicate better classification results.
It is obvious that the classification results of the Sample IV outperform the other three sample sets on the classification evaluation criteria (most likely due to the larger sample size). In addition, Fig. 6 shows the ROC curve of the classification results of different sample sets.
The AUC value of Sample IV is the largest among the four sample sets which indicate that the generated virtual samples can effectively improve the classification accuracy of the model.
Survival analysis
Kaplan–Meier survival curve [28] is the most commonly used method for survival analysis. It is an effective way to help us understand the occurrence of survival outcomes (endpoint events). The classification result of the two-branch DenseNet survival prediction model using the Sample IV is plotted as a Kaplan–Meier survival curve shown in Fig. 7.
The Kaplan–Meier survival curve shows that two-branch DenseNet survival prediction model is able to clearly divide patients into high-risk groups (red line in the figure) and low-risk groups (blue line in the figure), which exhibits good prediction ability and auxiliary effect on survival prediction of patients.
Discussion
Benefits of using two-branch architecture
Generally, one pathway is commonly used in the deep learning network. However, in this study, both T1 and T2 images are used to get more features for increasing the accuracy of classification. As we know, the two types of images have different or even opposite image features due to different imaging principles. Thus, a two-branch architecture is necessary for training T1 and T2 images separately, and then integrating all features to the classifier for survival prediction. Figure 8 shows the classification results using the single pathway model and the proposed two-branch model with different sample sets. Higher bars indicate better classification results. In order to know the comprehensive influence of the three variables, accuracy, sensitivity and specificity, on groups, they were taken as observation variables together, and the different model structures of the four groups of the legend were taken as control variables for the One-factor Analysis of Variance (ANOVA). The P value of significance test is 0.0189, indicating that the result is statistically significant and relatively reliable.
Figure 8 shows that the proposed two-branch DenseNet outperforms the single pathway model in accuracy, sensitivity and specificity regardless whether T1-weighted images, T2-weighted images, or mixed images are used.
Comparison with different feature extraction methods
In the two-branch model, the feature extraction method (network) used in each branch also affects the final classification result. The popular and common networks, CNN [29], VGG-16 [30], ResNet and DenseNet are compared in the framework of the two-branch model, and the accuracy, sensitivity and specificity are shown in Fig. 9. Higher bars indicate better classification results. It can be seen that the DenseNet outperforms other feature extraction networks in the classification and prediction of the survival of patients suffering from GBM. In order to know the comprehensive influence of the three variables, accuracy, sensitivity and specificity, on groups, they were taken as observation variables together, and the different feature extraction methods of the four groups of the legend were taken as control variables for the ANOVA. The P value of the significance test is 0.0334, indicating that the result is statistically significant and relatively reliable.
Benefits of using multi-channel features in classification
The three-channel samples used in this study combines three types of images: the whole slice T1 or T2 image, the image with tumor region only, and the image without tumor. The advantage of this method is that the diversity of features can be increased, and some features that are not available in single-channel samples such as the size and location of the tumor are retained. Figure 10 shows the classification results of using different channel samples. Higher bars indicate better classification results. In order to know the comprehensive influence of the three variables, accuracy, sensitivity and specificity, on groups, they were taken as observation variables together, and the different sample sets of the six groups of the legend were taken as control variables for the ANOVA. The P value of the significance test is 0.0179, indicating that the result is statistically significant and relatively reliable.
Figure 10 shows that using three-channel samples outperforms using single-channel and two-channel samples in the classification no matter which channels are chosen. In particular, we noticed that the sensitivity of the yellow bars is much lower and does not seem to follow the trend of the others. We speculated that because the yellow bar represents using the whole slice and the image without tumor as the two-channel sample for feature extraction, less attention is paid to the features of the most classification-relevant tumor region, that is, fewer features are extracted from tumor regions. As a result, the prediction results are biased toward one side, which increases the numerical gap between sensitivity and specificity.
Limitations and future works
In summary, this research has made some progress and proved the feasibility of the two-branch DenseNet survival prediction model, but some still remain to be improved. The two-branch architecture with multi-channel samples puts forward higher requirement to hardware for training the network, thus the number of the batch size usually need to be limited. These problems may lead to the insufficient training of the network. Additionally, the setting of the loss function is relatively important. For instance, focal loss function [31] has exhibited good performance in the case of unbalanced samples. However, we just deploy softmax loss functions which are commonly used in most architectures. So, it is necessary to pay more attention to selecting a more appropriate loss function for this study. With the deepening of the study, although we have improved the prediction result by using GANs, flipping and rotating methods to augment the sample size, the collection of more samples is still an important topic in the following work.
It's also worth pointing out that, the creation of artificial samples for survival prediction through GANs is unclear. The generated image may not be clinically consistent with the real image, but the purpose of introducing GANs this time is to expand the number of samples to achieve the goal of improving the accuracy of prediction classification, and the use of the expanded samples did achieve the predetermined goal. The image synthesis method used in this study does not involve the principles of biology and imaging, and only relies on the style transfer of deep learning methods to generate virtual images. At present, it is not suitable for image reconstruction and pathological research, but only suitable for sample expansion for other studies. It is well-known that deep learning-based neural networks provide far superior predictive power, but at the price of behaving as a "black-box" where the underlying reasoning is much more difficult to extract [32]. In order to verify the agreement between the AI and decision structure and their own ground-truth knowledge, explainable AI has developed as a subfield of AI, focused on exposing complex AI models to humans in a systematic and interpretable manner. So, explainable AI also should be focused in our future study, which is crucial to limit the possibility of improper, non-robust, and unsafe decisions and actions.
Conclusion
In this study, to compensate for the shortcomings of the previous survival prediction methods based on radiomics and machine learning, such as the manual feature extraction and screening and the lack of medical images, an end-to-end two-branch DenseNet survival prediction model relying on the feature extraction and screening ability of the deep network itself is proposed to predict the survival of patients suffering from GBM. Moreover, the one2one CycleGAN method is used to generate virtual samples, which can alleviate the situation of insufficient data and has a positive effect on tumor feature extraction. Instead of using the traditional single-channel medical images, three-channel samples are used in the experiment. The experiment results show that the two-branch DenseNet survival prediction model using the three-channel augmented original and virtual sample sets can achieve a classification accuracy of up to 94%, which demonstrates that the model is worth applying in the field of survival prediction of patients suffering from GBM. Still, it can be further improved to obtain greater practicability.
References
Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, Stroup N, Kruchko C, Barnholtz-Sloan J (2014) CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro-Oncology 16:iv1–iv63. https://doi.org/10.1093/neuonc/nos218
Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C (2016) Barnholtz-Sloan JS (2016) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro-oncology 18:v1–v75. https://doi.org/10.1093/neuonc/now207
Kenneth C, Bruce V, Kirk S, John F, Justin K, Paul K, Stephen M, Stanley P, David M, Michael P, Lawrence T, Fred P (2013) The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26(6):1045–1057. https://doi.org/10.1007/s10278-013-9622-7
Chang K, Zhang B, Guo X, Zong M, Rahman R, Sanchez D, Winder N, Reardon DA, Zhao B, Wen PY, Huang RY (2016) Multimodal imaging patterns predict survival in recurrent glioblastoma patients treated with bevacizumab. Neuro-oncology 18(12):1680–1687. https://doi.org/10.1093/neuonc/now086
Osman AFI (2018) Automated brain tumor segmentation on magnetic resonance images and patients overall survival prediction using support vector machines. BrainLes 2017 10670:435–449. https://doi.org/10.1007/978-3-319-75238-9_37
Ahmad C, Christian D, Matthew T, Bassam A (2017) Predicting survival time of lung cancer patients using radiomic analysis. Oncotarget 8(61):104393–104407. https://doi.org/10.18632/oncotarget.22251
Xue Y, Xu T, Zhang H, Long LR, Huang X (2018) SegAN: adversarial network with multi-scale L1 loss for medical image segmentation. Neuroinformatics 16(3/4):383–392. https://doi.org/10.1007/s12021-018-9377-x
Kumar A, Kim J, Lyndon D, Fulham M, Feng D (2017) An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J Biomed Health Inform 21(1):31–40. https://doi.org/10.1109/JBHI.2016.2635663
Sun D, Wang M, Li A (2018) A multimodal deep neural network for human breast cancer prognosis prediction by integrating multi-dimensional data. IEEE/ACM Trans Comput Biol Bioinf. https://doi.org/10.1109/TCBB.2018.2806438
Bello GA, Dawes TJW, Duan J, Carlo B, de Marvao A, Howard LSGE, Gibbs JSR, Wilkins MR, Cook SA, Daniel R, O’Regan DP (2019) Deep learning cardiac motion analysis for human survival prediction. Nat Mach Intell. https://doi.org/10.1038/s42256-019-0019-2
van der Burgh HK, Schmidt R, Westeneng HJ, de Reus MA, van den Berg LH, van den Heuvel MP (2017) Deep learning predictions of survival based on MRI in amyotrophic lateral sclerosis. NeuroImage Clin 13:361–369. https://doi.org/10.1016/j.nicl.2016.10.008
Nie D, Lu J, Zhang H, Ehsan A, Wang J, Yu Z, Liu LY, Wang Q, Wu J, Shen D (2019) Multi-channel 3D deep feature learning for survival time prediction of brain tumor patients using multi-modal neuroimages. Sci Rep 9:1103. https://doi.org/10.1038/s41598-018-37387-9
Tabibu S, Vinod PK, Jawahar CV (2019) Pan-renal cell carcinoma classification and survival prediction from histopathology images using deep learning. Sci Rep. https://doi.org/10.1038/s41598-019-46718-3
Gao H, Zhuang L, van der Maaten L, Weinberger KQ (2016) Densely connected convolutional. Networks. https://doi.org/10.1109/CVPR.2017.243
Shen Z, Zhou SK, Chen Y, Georgescu B, Liu X, Huang TS (2019) One-to-one mapping for unpaired image-to-image. Translation. https://doi.org/10.1109/WACV45572.2020.9093622
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2014) Generative adversarial nets. Adv Neural Inf Process Syst. https://doi.org/10.1007/978-1-4842-3679-6_8
Zhu JY, Park T, Isola P, Efros AA (2017) Unpaired image-to-image translation using cycle-consistent adversarial networks. https://doi.org/10.1109/ICCV.2017.244
Isola P, Zhu JY, Zhou T, Efros AA (2017) Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. https://doi.org/10.1109/CVPR.2017.632
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. Comput Vis Pattern Recognit. https://doi.org/10.1109/CVPR.2016.90
Bloice MD, Stocker C, Holzinger A (2017) Augmentor: an image augmentation library for machine learning. J Open Source Softw 2(19):5–6. https://doi.org/10.21105/joss.00432
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber M-A, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp C, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P (2015) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34(10):1993–2024. https://doi.org/10.1109/TMI.2014.2377694
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, Davatzikos C (2017) Advancing the Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Nat Sci Data 4:170117. https://doi.org/10.1038/sdata.2017.117
Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, Shinohara R, Berger C, Ha S, Rozycki M, Prastawa M, Alberts E, Lipkova J, Freymann J, Kirby J, Bilello M, Fathallah-Shaykh H, Wiest R, Kirschke J (2018) Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. https://doi.org/10.17863/CAM.38755
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP (2004) Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 13(4):600–612. https://doi.org/10.1109/TIP.2003.819861
Godbole S, Sarawagi S (2004) Discriminative methods for multi-labeled classification. In: 8th Pacific/Asia conference on advances in knowledge discovery and data. https://doi.org/10.1007/978-3-540-24775-3_5
Fawcett T (2006) An introduction to ROC analysis. Pattern Recognit Lett 27(8):861–874. https://doi.org/10.1016/j.patrec.2005.10.010
Hanley JA, Mcneil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143(1):29–36. https://doi.org/10.1148/radiology.143.1.7063747
Efron B (1988) Logistic regression, survival analysis, and the Kaplan-Meier curve. J Am Stat Assoc 83(402):414–425. https://doi.org/10.2307/2288857
Krizhevsky A, Sutskever I, Hinton G (2012) ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 25(2):1097–1105. https://doi.org/10.1145/3065386
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. Comput Sci. https://arxiv.org/abs/1409.1556
Lin T-Y, Goyal P, Girshick R, He K, Dollar P (2017) Focal loss for dense object detection. IEEE Trans Pattern Anal Mach Intell. https://doi.org/10.1109/ICCV.2017.324
Samek W, Montavon G, Vedaldi A, Hansen L, Müller K-R (2019) Explainable AI: Interpreting, Explaining And Visualizing deep. Learning. https://doi.org/10.1007/978-3-030-28954-6
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 61773205).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Fu, X., Chen, C. & Li, D. Survival prediction of patients suffering from glioblastoma based on two-branch DenseNet using multi-channel features. Int J CARS 16, 207–217 (2021). https://doi.org/10.1007/s11548-021-02313-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02313-4