Abstract
Purpose:
Augmented reality can improve the outcome of hepatic surgeries, assuming an accurate liver model is available to estimate the position of internal structures. While researchers have proposed patient-specific liver simulations, very few have addressed the question of boundary conditions. Resulting mainly from ligaments attached to the liver, they are not visible in preoperative images, yet play a key role in the computation of the deformation.
Method:
We propose to estimate both the location and stiffness of ligaments by using a combination of a statistical atlas, numerical simulation, and Bayesian inference. Ligaments are modeled as polynomial springs connected to a liver finite element model. They are initialized using an anatomical atlas and stiffness properties taken from the literature. These characteristics are then corrected using a reduced-order unscented Kalman filter based on observations taken from the laparoscopic image stream.
Results:
Our approach is evaluated using synthetic data and phantom data. By relying on a simplified representation of the ligaments to speed up computation times, it is not estimating the true characteristics of ligaments. However, results show that our estimation of the boundary conditions still improves the accuracy of the simulation by 75% when compared to typical methods involving Dirichlet boundary conditions.
Conclusion:
By estimating patient-specific boundary conditions, using tracked liver motion from RGB-D data, our approach significantly improves the accuracy of the liver model. The method inherently handles noisy observations, a substantial feature in the context of augmented reality.
Similar content being viewed by others
Introduction
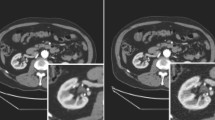
Primary liver cancer is one of the most common cancers in Europe and the fifth most common cause of cancer mortality [1]. Although several therapies are available to treat hepatic cancers, tumor resection remains the most efficient solution for patients with intrahepatic cholangiocarcinoma and, in some cases, patients with hepatocellular carcinoma [2]. The central function of the liver and its complex anatomy make it, however, challenging to remove these tumors, in particular, the small ones. A possible solution for guiding surgeons during these procedures is to use an augmented reality (AR) system. By fusing preoperative data with intraoperative images, and tracking liver motion over time, it is possible to overlay the internal structures of the organ onto the live view of the operating field (Fig. 1).
The liver is a soft organ, which changes in shape during surgery but also due to respiratory and cardiac motion. Providing an augmented view of the liver during surgery therefore requires to solve several challenges: the 3D model of the virtual anatomy needs to be registered onto the real organ using only partial surface data; it needs to follow actual tissue motion in real time; and it has to provide an estimate of the location of the internal structures with a good accuracy. To achieve these goals, different solutions were proposed, relying on different medical imaging modalities. The main approaches are either intensity-based methods, which attempt to fuse the preoperative data with the intraoperative image [3, 4] or, on the other side, physics-based methods, which describe, more or less accurately, the organ deformation [5, 6]. During hepatic surgery, the main visual system remains a direct vision (in open surgery) or through a camera (in laparoscopic procedures). As a result, only a small part of a liver surface is observed, from which a dense displacement field has to be recovered. In this context, a physically-based solution is very efficient at estimating the in-depth displacement from surface motion, or more generally from sparse data [7, 8].
In this context, the choice of a deformation model is obviously a key to obtaining an accurate, and fast, computation of the deformation. There are three main aspects to this: (1) the constitutive law that describes the stress–strain relationship, which in the case of the liver is known to be hyperelastic; (2) a numerical technique to solve this nonlinear partial differential equation on the domain of the organ, such as the finite element method; and (3) a set of material properties and boundary conditions (BCs) that characterize this particular organ. While an important body of work exists in the field of liver biomechanics [9, 10], fewer works have addressed the question of real-time computation [11] required for AR application. Yet, even fewer works have investigated the role of BCs in the computation of the soft tissue deformation.
The difficulty essentially comes from the impossibility to identify ligaments (and other anatomical structures constraining the liver) from the preoperative CT images (the dominant imaging modality for hepatic imaging). First, the description of liver connective tissues is limited to a generic idea about their location. Second, information about the biomechanical properties of ligaments is generally missing. To address the first issue, Plantefeve et al. [12] proposed a method for modeling boundary conditions in deformable anatomical structures, using a statistical atlas which gathers information about the connective structures attached to the organ. The atlas is then transferred to a specific patient’s anatomy using a physics-based registration technique, and the resulting boundary conditions are based on the mean position and variance of the ligaments. While this approach already shows a benefit in integrating this information in the model, it is not truly patient-specific nor does it model correctly these boundary conditions. As a result, it is nearly impossible to get a correct description of the liver boundary conditions.
In this paper, we propose to automatically identify and parameterize boundary conditions. Despite the fact that the liver motion is constrained by different anatomical structures, in this work we consider only the role of ligaments, which cover an important part of the liver and play an essential role in its behavior. The idea of identifying boundary conditions has been addressed by a few authors. In [13], the authors estimate what they call compliance boundary conditions. The authors consider an observed area as a separate object and suppose that the attachment between it and the outer part forms an additional constraint for the deformation, the properties of which could be estimated by comparing unconstrained and constrained motions. For this approach, in the case of a volumetric object, a sufficient amount of internal and accurate observations need to be obtained. In [14], the authors try to estimate BCs based on two deformed configurations of a liver. Matching the shapes gives a constraint that was applied to get this transformation. And based on this, it is possible to estimate the attached BCs. Unfortunately, two different shapes are necessary for this approach, whereas only one is usually taken for diagnosis purposes. In [15], an inverse simulation method is proposed. The authors present BCs as forces with unknown intensities. To find them, they apply traction and solve a gap minimization problem between simulated and observed positions. Here, it is implicitly supposed that the model is accurate enough to simulate the real object. But in the context of AR, the accuracy is often waived in favor of performance. The observed positions also have to be measured accurately.
In this work, we propose a solution to estimate liver boundary conditions in order to have a more accurate simulation of its deformation. It relies on the combination of a liver model and a nonlinear ligament simulation, initialized from a statistical atlas and corrected by a stochastic data assimilation process. Section 2 presents the details of the method, Sect. 3 shows our results on both synthetic and phantom data, and in Sect. 4, we discuss the benefits of our solution with regard to simulation accuracy.
Materials and methods
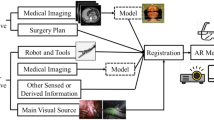
This section describes the main steps of our method. Section 2.1 presents our modeling choices, from which we derive optimized models in Sect. 2.3, while Sect. 2.2 details our data assimilation framework, based on an unscented Kalman filter. In our approach, we keep an important focus on patient-specific modeling and the possibility to deploy our method in a clinical context without the need for additional equipment. Figure 2 summarizes our approach.
Biomechanical modeling of the liver and its ligaments
Anatomy
The liver is the largest organ in the body, situated in the upper part of the abdomen on the right side [16]. The liver is connected to the lower part of the diaphragm and to the anterior wall of the abdomen by five ligaments: falciform, coronary, two triangular ligaments, and round ligament [17]. The coronary and triangular ligaments connect the liver to the diaphragm. Together with the connection of the inferior vena cava, they fix the posterior part of the liver. The falciform ligament does not give a lot of support, but it might limit the sideway displacement. The liver is also attached to the stomach by the hepatogastric ligament and to the duodenum by the hepatoduodenal ligament [16]. An overview of these ligaments is presented in Fig. 3.
The main boundary conditions for the liver are formed by the peritoneum, from which derive most of the ligaments (falciform, coronary, triangular, hepatogastric, and hepatoduodenal), while the round ligament is a fibrous cord resulting from the umbilical vein, and therefore has different properties. Below, we describe in more detail the biomechanics of the peritoneum and the ligaments it derives from, and look at possible constitutive models.
Biomechanics
Like many other soft tissues and organs in the human body, the liver has nonlinear behavior, which is presented in Fig. 4. Its average Young’s modulus is 10 kPa, and its Poisson’s ratio is close to 0.5. Various hyperelastic materials have been proposed to simulate its deformation: from Saint Venant Kirchhof (StVK) for real-time computation [18] up to porous visco-hyperelastic for accurate simulation [10]. Alternative solutions using a co-rotational model have been proposed [11] showing results similar to a StVK model. In this work, we consider the liver as a first order Ogden (hyperelastic) material. The relation between stress and strain is expressed through the strain energy density function \(\Psi \) as:
where \( \mu _1\) and \( \alpha _1\) are material parameters, and \( \lambda _i \) are principal stretches.
Regarding the biomechanics of the human peritoneum, a review of the literature does not provide any information about it. A few experiments were performed to study the mechanical properties of porcine [21] and bovine [22] peritoneum, but they give only a general idea. On the other hand, the properties of ligaments and tendons, which are also collagen-based connective tissues, are studied in more detail [23]. From an analysis performed by Witz et al. [24] showing that the peritoneum shares several similarities with ligaments, we choose to rely on the biomechanics of “true” ligaments for our work, which general stress–strain curve is presented in Fig. 4. Ligaments contain twisted fibers, which resistance for small strains is limited. As fibers are straightened, the stress increases rapidly, leading to a strong nonlinear stress–strain relation. As for the liver, various models were proposed to describe this nonlinear behavior: hyperelastic, quasi-linear viscoelastic, and nonlinear viscoelastic materials. In this work, we consider the ligaments as a Neo-Hookean material:
where \( \mu _1\) is the first Lame coefficient, characterizing material stiffness, and \(I_1\) is the trace of the Green deformation tensor.
Numerical simulation
Solving nonlinear partial differential equations for materials deformation, such as (2), requires an appropriate numerical technique. The finite element method (FEM) is often used in this case. It requires a discretization of the domain into simple geometrical shapes, which can be either surface or volume elements.
In this work, we model the liver using tetrahedral elements, which are easy to generate from a 3D surface mesh of the organ. This surface reconstruction is also almost automatic to compute from the segmented image of the liver. Ligaments, on the contrary, are very thin structures which are very challenging to segment, assuming they are visible in the preoperative image. Also, their geometry calls typically for a surface discretization using primitives such as triangles. Shells and membranes are usual options for modeling such structures while accounting for their small thickness. It is also possible to model such thin structures using volume elements. Regardless of this choice, it is obviously very difficult to construct an appropriate mesh for ligaments due to the lack of information about their shape. We address this point in the following sections.
Estimating boundary conditions using data assimilation
The variability of organ shapes and biomechanical characteristics across patients requires a method able to estimate BCs, as we cannot simply rely on an anatomical atlas. To deal with this uncertainty, we propose to use Bayesian inference to estimate both ligaments’ location and stiffness. In a nutshell, this approach uses Bayes’ theorem to update the probability for a hypothesis when more evidence or information becomes available. Bayesian inference takes into account the statistical noise on the data and provides a statistical regularization, which makes inverse problems with limited observations solvable. An efficient implementation of a Bayesian inference method able to process nonlinear systems like our models is the unscented Kalman filter (UKF) [25]. Compared to an extended Kalman filter, it does not require to compute the Jacobian of the system, which would be prohibitive given the size of our problem. The unknown data to be estimated (the stochastic state of the system) is described as a Gaussian distribution, which transformation through the nonlinear system is performed using an unscented transformation (see [25] for details). The main idea is to parameterize the Gaussian distribution using a set of sigma points, which hold the mean and covariance information, but are easier to transfer through a nonlinear function. The general algorithm is described in Algorithm 1. It consists of a loop that contains two main steps. During the prediction step, we form the new hypothesis about the estimated state, while during the correction step we correct it by comparing the predicted measurements with (noisy and partial) observations.

Each sigma point in the prediction step corresponds to perturbed parameter values and model positions (line 9 of the algorithm). The FE system is solved by taking the modified positions instead of original ones. Then, the predicted state is computed as a mean over FE simulation results for all sigma points. Therefore, the prediction step can be very costly when using a model with many degrees of freedom, as it is the case when using a FEM method. Using the simplex method to generate the sigma points, and with a mesh of N nodes and K stiffness parameters, this would mean \(3N+K+1\) simulations. A simple FEM mesh of only a few hundred nodes would be too time-consuming for a clinical application, as it would require more than 300 simulations for each step of the assimilation process. To solve this issue, we use a reduced-order unscented Kalman filter (ROUKF) [26] instead of the UKF. This method significantly reduces the computation cost since only \(K+1\) simulations (in the best case) are required. A ROUKF algorithm was also employed in [27]; however, the authors used a very simple model for the boundary conditions and did not rely on anatomical data to initialize the assimilation process.
Our approach being stochastic by design, it makes sense to initialize the location of the ligaments on the liver using a statistical atlas. Such an atlas is constructed from 15 manually annotated 3D liver reconstructions. On each liver, the estimated location of the ligaments has been identified by an expert surgeon, along with an uncertainty map. This allows to determine a probability distribution of the location of the ligaments on the surface of a human liver (see Fig. 5). The next step is to generate a discretization of the surface of the ligaments from these statistically relevant locations. Computing a finite element mesh, as generally done in biomechanics, would prove very difficult due to the non-deterministic nature of the ligament shape. It would also be computationally expensive to use in the parameter assimilation process. For these reasons, we propose an alternative solution in the following section.
The observations required to correct the model prediction and estimate the unknown parameters are typically obtained during surgery. Since it is difficult to observe the ligaments, we mainly rely on features tracked on the liver surface (using a variety of computer vision techniques). This requires to have a process model of the combined liver and ligament system. Only in that condition can we estimate BCs while observing the liver. Practically speaking, such intraoperative observations can be obtained using a stereoscopic laparoscope or an RGB-D camera (we need to have 3D observations).
Adapted model for assimilation process
Since our work is aimed at providing a predictive simulation in the context of augmented reality, we need to ensure that our modeling choices are compatible with this constraint. Similarly, the assimilation process allowing the estimation of the BCs needs to be as computationally efficient as possible. The ability to handle soft tissue cutting in the simulation is also of primary importance. The impact of such operations in terms of tissue modeling is considerable since it implies topological changes over time, the cost of which depends largely on the chosen geometrical representation but also on the numerical method that is adapted to compute tissue deformation. Here, we explain how we model the liver and its ligaments such that their simulation can be done in real time, and the data assimilation can be done efficiently.
Determination of a ligament location based on a set of available liver models where ligaments are labeled by an expert (white strips on the set of models on the left) on a new liver model (right model). A registered statistical atlas has expected ligament location (green strips) and a range of probable locations (white region).
To perform the numerical simulation of the liver, we rely on the finite element (FE) method with StVK material, a computationally efficient hyperelastic material often used for real-time simulations [18, 28]. To model ligaments, we use a discretization where the object is described as a set of mass points connected with springs, which are responsible for simulating the elastic property of the material. Besides being computationally efficient, meshing the region of the ligaments is easier. Yet, to represent the nonlinear stress–strain relationship of ligaments, we cannot rely on linear spring models, as usually proposed in the literature. To this end, we use a cubic spring, which internal force \( f\) is given by:
where \( k_\alpha \), \( k_\beta \), and \( k_\gamma \) are the stiffness parameters for a spring and \( \Delta l_{\mathrm{cur}} \) is the length variation. In addition, \(f= 0\) when \( \Delta l_{\mathrm{cur}} < 0\) to represent the fact that ligaments only generate forces on the liver under extension. To determine the values of \( k_\alpha \), \( k_\beta \), and \( k_\gamma \), we consider experimental data given in articles describing pig peritoneum mechanics [21] as well as human ligaments [29]. Based on this data, we modeled a thin rectangular shape (\(40\times 100\times 1\hbox { mm}^3\)) using a Neo-Hookean material and choose a parametrization that would produce similar stress–strain curves as [21] or [29]. This led to a set of constitutive laws with Young’s modulus ranging from 5 to 300 MPa. Then, we generated a discretization of the same domain using springs and assimilated the springs’ parameters. Since only the region of the ligament attached to the liver has an influence on its motion, we compare only the nodes in this region (marked as yellow spheres in Fig. 6). Results show an average error between both models of less than 2.4 mm in a stretching scenario, for strains up to 45% (i.e., covering the range of ligaments strain in surgery). This shows that the behavior of our mass–spring system is similar to a Neo-Hookean material. Yet it is 35 times faster to compute and can be automatically generated from the atlas.
Overview of the cubic spring model parametrization. To cover all possible ranges of ligaments behavior, maximal and minimal envelope curves are selected from given data (a), Neo-Hookean parameters are defined, and a FE simulation of a stretching experiment is computed (b (top)). Parameters for the mass–spring system are then assimilated from the FE simulation (b (bottom)). The average difference, measured for validation points (yellow markers), between strained model bounds for strains up to 45% remains small (right).
Experimental results
Numerical validation
We first validated our optimized liver model against a numerical solution. The liver was generated from a segmented CT scan of a patient, and the ligaments locations were defined by an expert. For simplicity, we decided to consider only the falciform ligament. The reference simulation was computed using FEBio [30]. The liver was meshed with about 73,000 linear tetrahedral elements. The ligament meshes (shell elements) were generated by extrusion in the direction of the liver surface normal. Dirichlet constraints were applied to the nodes furthest from the liver, and bilateral constraints used to couple the liver and ligament models. An Ogden model was used for the liver with parameters taken from [31] (\(\alpha _1=0.88\), \(\mu _1=16.47\) kPa, \(\text {G}=7.21\) kPa) Neo-Hookean material for ligaments (Young’s modulus of 100 kPa, Poisson’s ratio of 0.48). To generate a deformation representative of surgical manipulation, we applied non-constant periodic loads to both lobes of the liver, marked as F1, F2, and F3 in Fig. 7. We also defined 14 virtual markers that represent observations that could be obtained during an actual intervention. They are used in the data assimilation process.
Left: numerical simulation mimicking a manipulation of the liver, from which observations are extracted. Right: difference, computed at validation markers, between the numerical ground truth, a simulation using fixed BCs based on the anatomical atlas (green line), and our real-time simulation (green line), after estimation of the BCs.
On the other hand, the real-time liver and ligament model was implemented using SOFA [32]. The liver mesh was generated with approximately 7,500 elements. We used a StVK model, for which a Poisson’s ratio of 0.49 was set. The Young’s modulus was computed using data from [31]. The ligaments were also extruded along the liver surface normal as two layers of nonlinear springs. The spring parameters were inferred from the Neo-Hookean model in FEBio using the assimilation method described previously (ROUKF). To further optimize the assimilation process, we split the set of springs into groups that share the same stiffness parameters. This way, we estimate the parameters for each region, not for each spring. Again, this is just an optimization to speed up the estimation of the BCs and can be adjusted depending on the amount of data or time available.
To assess our results, we use virtual markers uniformly distributed inside the liver model. To estimate how our BC estimation can improve the overall accuracy of the liver deformation, we compare the marker positions obtained with our method with a simulation involving the same FEM liver model but using fixed BCs. The location of the BCs is, however, also based on the anatomical atlas, to avoid any bias in the comparison. The results show that for BCs estimated with our method, the mean error in the deformation is only 1.99 mm (\(\pm \,0.7~\hbox {mm}\)), while it is 7.6 mm (\(\pm \, 4.6~\hbox {mm}\)) when using predefined constant BCs. For the largest deformation, the error is reduced by 75%, as shown in Fig. 7.
Augmented reality on a liver phantom
To further demonstrate the benefit of our method, we applied it to phantom liver data. The liver phantom was made of silicone foam (Soma Foama 15 from Smooth-On, Inc.), which has elastic behavior similar to the actual liver, of half the size of an actual liver, and a piece of fabric made from polyester was used to mimic the falciform ligament. The model was placed inside a wooden frame, with a support for an Intel Realsense D435 RGB-D camera. Several passive retroreflective markers were placed on the visible part of a phantom. They were tracked using a pyramidal Lucas–Kanade algorithm [33], which together with depth data were used to obtain 3D observations. An overview of the setup is presented in Fig. 8. We then manipulate the liver similarly as in Sect. 3.1 and record the motion of a small subset of the point cloud generated from the depth map of the RGB-D camera. This is used to assimilate the parameters of the ligament model. Finally, the virtual liver model is overlaid onto the camera stream, and its deformation computed using the (now parametrized) model and the point cloud returned by the camera. An example of an AR view is presented in Fig. 8. The computation time for the combined liver and ligament models is 20 ms, allowing for a real-time update of the augmented view.
Conclusion
In this paper, we proposed a new method for improving the accuracy of a numerical simulation of the liver, by dynamically estimating boundary conditions. The assimilation method, based on ROUKF, uses observations obtained from laparoscopic or open surgery images. The liver and ligament models are simulated using a coupled system consisting of a real-time FEM and a cubic spring–mass system. The spring network describes the unknown BCs linking the liver to its environment. Once their location and stiffness parameters have been estimated by the filter, a more accurate patient-specific simulation of the liver behavior can be obtained. The gain over usual approaches is significant, with errors reduced by up to 10 mm in certain cases.
The main advantage of this approach is its stochastic nature. Therefore, first of all, it takes into account that intraoperative images have observation errors. Secondly, together with results, it gives us estimation statistics, which we could use for additional information, like exploring the observability of the system. The filtering approach also allows to get intermediate results, which could be used to improve the accuracy of the AR system in a workflow process.
However, the filtering process can be quite time-consuming, in particular, if we consider all ligaments connected to the liver since a simulation step has to be computed for every sigma point (and the number of sigma points is proportional to the number of unknowns to estimate). A possible solution to speed up the data assimilation process (and ideally reach real-time computation) is to use preconditioning. Although the cost of computing a preconditioner is relatively high, it might be possible to use the same preconditioner for all of the sigma points (depending on the sensibility of the system matrix to the parameters to estimate). This would reduce the computation time considerably.
References
Ferlay J, Parkin DM, Steliarova-Foucher E (2010) Estimates of cancer incidence and mortality in europe in 2008. Eur J Cancer 46(4):765–781
Orcutt ST, Anaya DA (2018) Liver resection and surgical strategies for management of primary liver cancer. J Moffitt Cancer Center 25(1):1073274817744621
Puerto-Souza GA, Mariottini GL (2013) Toward long-term and accurate augmented-reality display for minimally-invasive surgery. In: IEEE ICRA, pp 5384–5389
Chen L, Tang W, John NW (2017) Real-time geometry-aware augmented reality in minimally invasive surgery. Healthc Technol Lett 4(5):163–167
Haouchine N, Dequidt J, Peterlik I, Kerrien E, Berger M-O, Cotin S (2013) Image-guided simulation of heterogeneous tissue deformation for augmented reality during hepatic surgery. In: 2013 IEEE ISMAR, pp 199–208
Suwelack S (2015) Real-time Biomechanical Modeling for Intraoperative Soft Tissue Registration. Ph.D. thesis, Karlsruhe Institute of Technology
Peterlík I, Courtecuisse H, Rohling R, Abolmaesumi P, Nguan C, Cotin S, Salcudean S (2018) Fast elastic registration of soft tissues under large deformations. Med Image Anal 45:24–40
Clements LW, Collins JA, Weis JA, Simpson AL, Kingham TP, Jarnagin WR, Miga MI (2017) Deformation correction for image guided liver surgery: an intraoperative fidelity assessment. Surgery 162(3):537–547
Chui C-K, Kobayashi E, Chen X, Hisada T, Sakuma I (2007) Transversely isotropic properties of porcine liver tissue: experiments and constitutive modelling. Med Biol Eng Comput 45(1):99–106
Marchesseau S, Chatelin S, Delingette H (2017) Nonlinear biomechanical model of the liver. In: Biomechanics of living organs, vol 1, Elsevier, Amsterdam, pp 243–265
Peterlík I, Duriez C, Cotin S (2012) Modeling and real-time simulation of a vascularized liver tissue. In: MICCAI, pp. 50–57
Plantefève R, Peterlik I, Courtecuisse H, Trivisonne R, Radoux J-P, Cotin S (2014) Atlas-based transfer of boundary conditions for biomechanical simulation. In: MICCAI, pp 33–40
Ozkan E, Goksel O (2018) Compliance boundary conditions for patient-specific deformation simulation using the finite element method. Biomed Phys Eng 4(2):025003
Peterlik I, Courtecuisse H, Duriez C, Cotin S (2014) Model-based identification of anatomical boundary conditions in living tissues. In: IPCAI, pp 196–205
Largillière F, Coevoet E, Sanz-Lopez M, Grisoni L, Duriez C (2016) Stiffness rendering on soft tangible devices controlled through inverse fem simulation. In: Proceedings of IROS, pp 5224–5229
Gray H (1918) Anatomy of the human body. Lea & Febiger, Philadelphia
Abdel-Misih SRZ, Bloomston M (2010) Liver anatomy. Surg Clin N Am 90(4):643–653
Delingette H, Ayache N (2004) Soft tissue modeling for surgery simulation. In: Computational models for the human body, vol 12, Elsevier, Amsterdam, pp 453–550
Nava A, Mazza E, Kleinermann F, Avis NJ, McClure J, Bajka M (2004) Evaluation of the mechanical properties of human liver and kidney through aspiration experiments. Technol Health Care 12(3):269–280
Rodriguez Lelis J, Médez Aguirre J, Arellano Cabrera J, Navarro Torres J, Pliego A (2011) On the mechanical design of cruciate ligaments based on polymeric ropes for knee prosthesis. Revista Mexicana de Ingenieria biomédica 32(1):25–31
White EJ, Cunnane EM, McMahon M, Walsh MT, Coffey JC, O’Sullivan L (2018) Mechanical characterisation of porcine non-intestinal colorectal tissues for innovation in surgical instrument design. J Eng Med 232(8):796–806
Sarac TP, Carnevale K, Smedira N, Tanquilut E, Augustinos P, Patel A, Naska T, Clair D, Ouriel K (2005) In vivo and mechanical properties of peritoneum/fascia as a novel arterial substitute. J Vasc Surg 41(3):490–497
Reese SP, Ellis BJ, Weiss JA (2013) Multiscale modeling of ligaments and tendons. Springer, New York, pp 103–147
Witz CA, Montoya-Rodriguez IA, Cho S, Centonze VE, Bonewald LF, Schenken RS (2001) Composition of the extracellular matrix of the peritoneum. J Soc Gynecol Invest 8(5):299–304
Julier SJ, Uhlmann JK, Durrant-Whyte HF (1995) A new approach for filtering nonlinear systems. In: Proceedings of ACC’95, vol 3, pp 1628–1632
Julier SJ, Uhlmann JK (2002) Reduced sigma point filters for the propagation of means and covariances through nonlinear transformations. In: Proceedings of the American Control Conference, vol 2, pp 887–892. https://doi.org/10.1109/ACC.2002.1023128
Peterlik I, Haouchine N, Ručka L, Cotin S (2017) Image-driven stochastic identification of boundary conditions for predictive simulation. In: MICCAI, pp 548–556
Barbič J, James D (2005) Real-time subspace integration for st. venant-kirchhoff defromable models. ACM Trans Graph 24(3):982–990
Forestiero A, Carniel EL, Natali AN (2014) Biomechanical behaviour of ankle ligaments: constitutive formulation and numerical modelling. Comput Methods Biomech Biomed Eng 17(4):395–404
Maas SA, Ellis BJ, Ateshian GA, Weiss JA (2012) Febio: finite elements for biomechanics. J Biomech Eng 134(1)
Lu Y-C, Kemper A, Gayzik S, Untaroiu C, Beillas P (2013) Statistical modeling of human liver incorporating the variations in shape, size, and material properties. In: 57th Stapp car crash conference, vol 57, pp 285–311
Allard J, Cotin S, Faure F, Bensoussan P-J, Poyer F, Duriez C, Delingette H, Grisoni L (2007) SOFA: an open source framework for medical simulation, vol 125. IOS Press, New York, pp 13–18
Bouguet J-Y (1999) Pyramidal implementation of the lucas kanade feature tracker. Intel Corporation, Microprocessor Research Labs, pp 1–10
Funding
This study was funded from the EU H2020 Research and Innovation programme under Marie Sklodowska-Curie Grant Agreement No. 722068.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
This articles does not contain patient data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nikolaev, S., Cotin, S. Estimation of boundary conditions for patient-specific liver simulation during augmented surgery. Int J CARS 15, 1107–1115 (2020). https://doi.org/10.1007/s11548-020-02188-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-020-02188-x