Abstract
Purpose
Morphometry techniques were applied to quantify the normal tissue therapy response in patients receiving whole-brain radiation for intracranial malignancies.
Methods
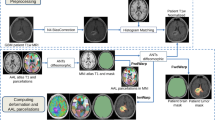
Pre- and Post-irradiation magnetic resonance imaging (MRI) data sets were retrospectively analyzed in N = 15 patients. Volume changes with respect to pre-irradiation were quantitatively measured in the cerebrum and ventricles. Measurements were correlated with the time interval from irradiation. Criteria for inclusion included craniospinal irradiation, pre-irradiation MRI, at least one follow-up MRI, and no disease progression. The brain on each image was segmented to remove the skull and registered to the initial pre-treatment scan. Average volume changes were measured using morphometry analysis of the deformation Jacobian and direct template registration-based segmentation of brain structures.
Results
An average cerebral volume atrophy of \(-\)0.2 and \(-\)3 % was measured for the deformation morphometry and direct segmentation methods, respectively. An average ventricle volume dilation of 21 and 20 % was measured for the deformation morphometry and direct segmentation methods, respectively.
Conclusion
The presented study has developed an image processing pipeline for morphometric monitoring of brain tissue volume changes as a response to radiation therapy. Results indicate that quantitative morphometric monitoring is feasible and may provide additional information in assessing response.





Similar content being viewed by others
Notes
ANTS -s CC[${fixed}${moving},1,5] -t SyN[0.25] -r Gauss[3,0] -i 30x90x20 –use-Histogram-Matching–number-of-affine-iterations 10000x10000x10000x10000x10000 –MI-option 32x16000.
References
Armstrong C, Ruffer J, Corn B, DeVries K, Mollman J (1995) Biphasic patterns of memory deficits following moderate-dose partial-brain irradiation: neuropsychologic outcome and proposed mechanisms. J Clin Oncol 13(9):2263–2271
Asai A, Matsutani M, Kohno T, Nakamura O, Tanaka H, Fujimaki T, Funada N, Matsuda T, Nagata K, Takakura K (1989) Subacute brain atrophy after radiation therapy for malignant brain tumor. Cancer 63(10):1962–1974
Avants BB, Epstein CL, Grossman M, Gee JC (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12(1):26–41. doi:10.1016/j.media.2007.06.004
Avants BB, Tustison NJ, Song G (2011) A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54(3):44–2033. doi:10.1016/j.neuroimage.2010.09.025
Avants BB, Yushkevich P, Pluta J, Minkoff D, Korczykowski M, Detre J, Gee JC (2010) The optimal template effect in hippocampus studies of diseased populations. Neuroimage 49(3):2457–2466
Bayouth JE, Casavant TL, Graham MM, Sonka M, Muruganandham M, Buatti JM (2011) Image-based biomarkers in clinical practice. Semin Radiat Oncol 21(2):157–166. doi:10.1016/j.semradonc.2010.11.003
Beg MF, Miller MI, Trouvé A, Younes L (2005) Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vis 61(2):139–157
Bro-Nielsen M, Gramkow C (1996) Fast fluid registration of medical images. Proc Vis Biomed Comput 4:267
Brown GG, Lee JS, Strigo IA, Caligiuri MP, Meloy M, Lohr J (2011) Voxel-based morphometry of patients with schizophrenia or bipolar i disorder: a matched control study. Psychiatry Res Neuroimaging 194(2):149–156
Castillo E, Castillo R, Fuentes D, Guerrero T (2014) Computing global minimizers to a constrained B-spline image registration problem from optimal l1 perturbations to block match data. Med Phys 41(4):041, 904
Castillo R, Castillo E, Fuentes D, Ahmad M, Wood AM, Ludwig MS, Guerrero T (2013) A reference dataset for deformable image registration spatial accuracy evaluation using the COPDgene study archive. Phys Med Biol 58(9):2861–2877
Castillo R, Castillo E, Guerra R, Johnson VE, McPhail T, Garg AK, Guerrero T (2009) A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets. Phys Med Biol 54(7):1849
Cherrier MM, Anderson K, David D, Higano CS, Gray H, Church A, Willis SL (2013) A randomized trial of cognitive rehabilitation in cancer survivors. Life Sci 93(17):617–622. doi:10.1016/j.lfs.2013.08.011
Christensen GE, Rabbitt RD, Miller MI (1996) Deformable templates using large deformation kinematics. IEEE Trans Image Process 5(10):1435–1447
DeAngelis LM, Delattre JY, Posner JB (1989) Radiation-induced dementia in patients cured of brain metastases. Neurology 39(6):789–789
Dupuis P, Grenander U, Miller MI (1998) Variational problems on flows of diffeomorphisms for image matching. Q Appl Math 56(3):587
Greene-Schloesser D, Robbins ME, Peiffer AM, Shaw EG, Wheeler KT, Chan MD (2012) Radiation-induced brain injury: a review. Frontiers Oncol 2(July):73. doi:10.3389/fonc.2012.00073
Holden M, Hill DLG, Denton ERE, Jarosz JM, Cox TCS, Rohlfing T, Goodey J, Hawkes DJ (2000) Voxel similarity measures for 3-D serial MR brain image registration. IEEE Trans Med Imaging 19(2):94–102
Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM (2012) Fsl. NeuroImage 62(2):782–790
Kalkanis SN, Linskey ME (2010) Evidence-based clinical practice parameter guidelines for the treatment of patients with metastatic brain tumors: introduction. J Neuro-Oncol 96(1):7–10
Klein AEA (2009) Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage 46(3):786–802
Lehmann M, Crutch SJ, Ridgway GR, Ridha BH, Barnes J, Warrington EK, Rossor MN, Fox NC (2011) Cortical thickness and voxel-based morphometry in posterior cortical atrophy and typical alzheimer’s disease. Neurobiol Aging 32(8):1466–1476
Liu AK, Marcus KJ, Fischl B, Grant PE, Young Poussaint T, Rivkin MJ, Davis P, Tarbell NJ, Yock TI (2007) Changes in cerebral cortex of children treated for medulloblastoma. Int J Radiat Oncol Biol Phys 68(4):992–998
Merchant TE, Hua CH, Shukla H, Ying X, Nill S, Oelfke U (2008) Proton versus photon radiotherapy for common pediatric brain tumors: comparison of models of dose characteristics and their relationship to cognitive function. Pediatr Blood Cancer 51(1):110–117
Mulhern RK, Palmer SL, Reddick WE, Glass JO, Kun LE, Taylor J, Langston J, Gajjar A (2001) Risks of young age for selected neurocognitive deficits in medulloblastoma are associated with white matter loss. J Clin Oncol 19(2):472–479
Murphy K, Van Ginneken B, Reinhardt JM, Kabus S, Ding K, Deng X, Cao K, Du K, Christensen GE, Garcia V et al (2011) Evaluation of registration methods on thoracic CT: the EMPIRE10 challenge. IEEE Trans Med Imaging 30(11):1901–1920
National Cancer Institute: A snapshot of: Brain and central nervous system cancers (2013)
Nieder C, Leicht A, Motaref B, Nestle U, Niewald M, Schnabel K (1999) Late radiation toxicity after whole brain radiotherapy: the influence of antiepileptic drugs. Am J Clin Oncol 22(6):573–579
Patchell RA, Tibbs PA, Regine WF, Dempsey RJ, Mohiuddin M, Kryscio RJ, Markesbery WR, Foon KA, Young B (1998) Postoperative radiotherapy in the treatment of single metastases to the brain. J Am Med Assoc 280(17):1485–1489
R Core Team: R (2013) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Reddick WE, Russell JM, Glass JO, Xiong X, Mulhern RK, Langston JW, Merchant TE, Kun LE, Gajjar A (2000) Magn Reson Imaging 18(7):787–793
Tanner JM (1962) Growth at adolescence. Thomas, Springfield, IL
Thirion JP (1995) Fast non-rigid matching of 3D medical image. Tech. rep., Research Report RR-2547, Epidure Project, INRIA Sophia
Thompson PM, Hayashi KM, Sowell ER, Gogtay N, Giedd JN, Rapoport JL, de Zubicaray GI, Janke AL, Rose SE, Semple J et al (2004) Mapping cortical change in alzheimer’s disease, brain development, and schizophrenia. Neuroimage 23:S2–S18
Torres I, Mundt A, Sweeney P, Llanes-Macy S, Dunaway L, Castillo M, Macdonald R (2003) A longitudinal neuropsychological study of partial brain radiation in adults with brain tumors. Neurology 60(7):1113–1118
Tustison NJ, Avants BB (2013) Explicit B-spline regularization in diffeomorphic image registration. Front Neuroinformatics 7:39
Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G (2006) User-guided 3d active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3):1116–1128
Zhang Y, Zou P, Mulhern RK, Butler RW, Laningham FH, Ogg RJ (2008) Brain structural abnormalities in survivors of pediatric posterior fossa brain tumors: a voxel-based morphometry study using free-form deformation. NeuroImage 42(1):218–29. doi:10.1016/j.neuroimage.2008.04.181. URL: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2591023&tool=pmcentrez&rendertype=abstract
Zou K, Wells W III, Kikinis R, Warfield S (2004) Three validation metrics for automated probabilistic image segmentation of brain tumours. Stat Med 23(8):1259–1282
Acknowledgments
This work is supported in part by the O’Donnell Foundation and NIH DP2OD007044, NIH DP2OD007044-01S1, and CPRIT RP101502 funding mechanisms. The authors would also like to thank the open source communities ITK, ANTs [4], itk-SNAP [37], and FSL [19] for providing enabling software for image processing and visualization.
Conflict of interest
The authors have no conflicts of interest to report.
Author information
Authors and Affiliations
Corresponding author
Appendix: Gaussian approximation of the Green’s Kernel
Appendix: Gaussian approximation of the Green’s Kernel
Following a diffeomorphic image registration framework [4, 7, 14], consider two images, \(I_0\) and \(I_1\), defined on an Eulerian reference domain, \(\varOmega \subset \mathbb {R}^3\). The goal of the image registration is to determine the motion, \(\mathbf {\varphi }(\varvec{x},t): \varOmega \times [0,1] \rightarrow \varOmega \), that maps the reference image, \(I_0\), to the current image, \(I_1\).
A symmetric diffeomorphic registration is optimal with respect to a given image similarity metric, \(d: \varOmega \times \varOmega \rightarrow \mathbb {R}\), and penalized by the velocity of the transformation.
Here, the deformations, \(\mathbf {\varphi }_i, i=1,2\), are defined with respect to the midpoint, \(t=.5\), of the transformation. Time is parameterized in opposite directions between \(\mathbf {\varphi }_1\) and \(\mathbf {\varphi }_2\). The operator norm \(\Vert \cdot \Vert _L\) is induced by a differential operator of the type, \(L=\alpha \varDelta + Id\), \(\alpha \in \mathbb {R}\) [7]. The symmetric diffeomorphic formulation mappings are constructed sufficiently smooth such that the inverse of the motion is well defined, \(\mathbf {\varphi }^{-1}(\varvec{x},t): \varOmega \times [0,1] \rightarrow \varOmega \), and gives a consistent solution for the forward and inverse mapping, \(\mathbf {\varphi }\circ \mathbf {\varphi }^{-1} = Id\).
The Euler–Lagrange equations provide necessary conditions for which a solution of the optimization formulation (3) must satisfy.
Here, the gradient of the objective function (3) is with respect to the velocity of the transformation. The operator \(L\) represents a physics constraint on the deformation solution. Assuming the deformation behaves as a viscous fluid provides an intuitive solution field that may be understood to adhere to first principle conservation laws.
Further, compared with linear elastic displacement models that constrain the accuracy of large deformations because of internal elastic strain, accurate large deformations may be achieved within this viscous fluid model because internal forces disappear over time and the desired deformation can be fully achieved [8]. However, this approach leads to computationally expensive numerical solution schemes that couple the individual components of the deformation. Alternatively, assuming each deformation component is decoupled and diffuses along the respective gradient of the deformation field yields the algorithmically and numerically tractable Gaussian convolution kernel used in SyN [4]. The greedy update at the midpoint in time simplifies to a fixed point iteration on the velocity field [36], and the transformation field is iteratively updated through a finite difference approximation of the velocity
Here, \(K \star \) represents the Gaussian convolution operation and is obtained from a Fourier transform solution of the heat equation. Solutions of the Euler–Lagrange equations may be interpreted as a decoupled component-wise solution to a isotropic heat transfer equation with initial conditions given by the gradient of the similarity metric.
Rights and permissions
About this article
Cite this article
Fuentes, D., Contreras, J., Yu, J. et al. Morphometry-based measurements of the structural response to whole-brain radiation. Int J CARS 10, 393–401 (2015). https://doi.org/10.1007/s11548-014-1128-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-014-1128-3