Abstract
Purpose
Aim of this study was to develop a generalised radiomics model for predicting pathological complete response after neoadjuvant chemo-radiotherapy in locally advanced rectal cancer patients using pre-CRT T2-weighted images acquired at a 1.5 T and a 3 T scanner.
Methods
In two institutions, 195 patients were scanned: 136 patients were scanned on a 1.5 T MR scanner, 59 patients on a 3 T MR scanner. Gross tumour volumes were delineated on the MR images and 496 radiomic features were extracted, applying the intensity-based (IB) filter. Features were standardised with Z-score normalisation and an initial feature selection was carried out using Wilcoxon–Mann–Whitney test: The most significant features at 1.5 T and 3 T were selected as main features. Several logistic regression models combining the main features with a third one selected by those resulting significant were elaborated and evaluated in terms of area under curve (AUC). A tenfold cross-validation was repeated 300 times to evaluate the model robustness.
Results
Three features were selected: maximum fractal dimension with IB = 0–50, energy and grey-level non-uniformity calculated on the run-length matrix with IB = 0–50. The AUC of the model applied to the whole dataset after cross-validation was 0.72, while values of 0.70 and 0.83 were obtained when 1.5 T and 3 T patients were considered, respectively.
Conclusions
The model elaborated showed good performance, even when data from patients scanned on 1.5 T and 3 T were merged. This shows that magnetic field intensity variability can be overcome by means of selecting appropriate image features.
Similar content being viewed by others
Introduction
Rectal cancer accounts for one third of colorectal cancers and is to date one of the leading causes of cancer death in the western world [1, 2].
Neoadjuvant chemo-radiotherapy (nCRT) followed by total mesorectal excision (TME) represents the standard of care for patients affected by locally advanced rectal cancer (LARC), defined as stage II (T3 or T4, node-negative, M0) and stage III (T3 or T4, node-positive, M0) rectal cancer.
In particular, nCRT reduces the risk of local recurrence and downsizes the primary tumour facilitating the subsequent successful surgical resection or allowing sphincter-preserving approaches. In a variable fraction of cases (8–30%), the downsizing effect is complete and a pathological complete response (pCR) after nCRT is reached [3,4,5].
The response after nCRT and outcomes are strongly correlated, as assessed in several pooled analyses, where complete responders show a better prognosis in terms of local control and disease free survival, compared to non-responders [6,7,8].
New therapeutic approaches, more conservative respect to the TME surgery, have currently under investigation for patients showed pCR after nCRT, such as local excision (LE) or watch and wait (W&W) [9,10,11].
To increase the number of locally advanced rectal cancer (LARC) patients with organ sparing treatment approaches, there is a growing interest in realising predictive models able to identify patients who will completely respond to nCRT before the start of therapy. These predictive models can be based on the analysis of clinical parameters, DNA sequences or radiomic parameters extracted by diagnostic images [12, 13].
An increasing number of studies focused on the possibility to predict pCR analysing magnetic resonance imaging (MRI), as this modality is generally the gold standard diagnostic imaging technique for rectal cancer [14, 15].
Different radiomics experiences performed on staging pre nCRT MRI have demonstrated that it is possible to predict complete response to treatment or early disease progression, relapse or distant progression in the first 3 months after radical surgery [16,17,18,19].
However, the application of radiomics to MRI is performed less often compared to radiomics on CT or positron emission tomography, due to the high complexity of the MRI signal processing, that require dedicated filtering and signal intensity standardisation procedures [20,21,22].
Another factor that obstructs the broad implementation of radiomics in daily clinical practice is the lack of evidence for the general applicability of the models for different MR scanner vendors and different magnetic field strengths [23].
Most of the imaging-based predictive models to date available are indeed strongly linked to the technical aspects of the used MR scanner [23, 24]. Aim of this study is to overcome these sources of variability, developing a radiomics model able to predict pCR in LARC patients analysing pre-CRT MR images acquired using MR scanners of different vendors and characterised by different magnetic field intensities (1.5 and 3 T).
Methods
Patients
A total of 195 LARC patients treated in two different medical centres (XXX, and YYY) were enrolled.
The cohort coming from Institution A (136 cases) was retrospectively enrolled among patients treated between May 2008 and December 2014; the cohort coming from Institution B (59 cases) considered patients among those treated between November 2008 and March 2012.
The inclusion criteria of this study were: patients affected by biopsy proven LARC with no evidence of distant metastases at staging radiological exams (Chest-Abdomen contrast enhanced CT and pelvic MRI); age of 18 years or more at the time of diagnosis.
No differences based on gender, age or ethnic group were considered for patient selection purposes. Informed consent was acquired from all the patients according the two ethical committees’ policies.
Treatment workflow and response assessment
All the patients were treated with to the same protocol: neoadjuvant long course chemo-radiotherapy followed by total mesorectal excision 6–8 weeks after the end of nCRT. The 25 fractions radiotherapy treatment was administered using a simultaneous integrated boost technique (55 Gy in fractions of 2.2 Gy to Gross Tumour Volume (GTV) and corresponding mesorectum and 45 Gy in fractions of 1.8 Gy to whole mesorectum and selected lymphatic drainage stations, according to disease stage) or a technique without a boost (50 Gy in fractions of 2.0 Gy to GTV, mesorectum and elective lymph node stations). Concomitant chemotherapy with chronomodulate Capecitabine (1650 mg/mq) or intravenous 5-Fluorouracil (5-FU) or an intensified schedule with Capecitabine (1300 mg/mq) plus Oxaliplatin (60 mg/mq) was prescribed, depending on the clinical stage and general conditions of the patient.
The pathological reports included histology, grading and tumour regression grade (TRG) according to Mandard classification [25]. Pathological complete response was defined as ypT0N0 or ypT0/ypNx.
Image analysis
The MR images were acquired using a protocol with T2-weighted images acquired in the plane orthogonal to the tumour longitudinal axis. The data were acquired using a 1.5 T MR scanner (GE Signa Exite, Little Chalfont, United Kingdom) in institution A, and a 3 T MR scanner (Philips Medical System, Eindhoven, The Netherlands) in institution B. No intravenous contrast agents were administered for both cohorts of patients.
The images had a spatial resolution of 0.703 × 0.703 mm2 for 1.5 T and 0.45 × 0.45 mm2 for 3 T. Slice thickness was 3 mm for 1.5 T and 3.5 mm for 3 T MR images.
The field of view was 18 cm for 1.5 T and 14 cm for 3 T MR images.
MR images were imported in a radiotherapy workstation (Eclipse, Varian Medical System™, Palo Alto, California, US) and the GTV was contoured by two experienced radiation oncologists, using the ICRU n.83 guidelines [26].
The DICOM files were imported in Moddicom, an R package (R Core Team, Vienna, Austria) designed for radiomics analyses of biomedical images [27, 28]. All the images were resampled to a spatial planar resolution of 0.7 × 0.7 mm2 prior to their quantitative analysis.
Feature extraction
A total of 90 radiomic features belonging to four families (fractal, statistical, textural and morphological features) were extracted from the images. Two image filters have been applied to the MR image before the feature extraction.
The intensity-based (IB) image filter proposed by Cusumano et al. was applied to the MR images before to extract radiomics features [22]. This approach consisted of normalising the signal intensity of the pixels inside the region of interest (ROI) analysed using as extremes the first and 99-th percentile of the grey-levels histogram representing the ROI. Pixel clusters were then identified considering two threshold levels (lower and upper level) defined as percentages of the maximum intensity level. All the images were filtered considered the IB filter with all the possible combinations of thresholds “lower level–upper level” for levels ranging from 0 to 100% by 10% (55 steps).
Considering the application of all the filters used, a total of 496 parameters were assessed.
Data analysis
A database was created combining the image features with clinical parameters (sex, age, clinical TNM staging) and outcome data (complete or not pathological response). Before merging the two cohorts of patients to a single training set, the homogeneity between the two datasets in terms of clinical parameters and pCR rates was assessed. The homogeneity was estimated using the Wilcoxon–Mann–Whitney (WMW) and Pearson’s χ2 test, depending on the nature of the variable investigated (WMW for continue variables, χ2 for categorical ones) [29].
Features were standardised with Z-score normalisation before features selection. Both cohorts were considered as training set for data analysis purposes, using cross-validation methods to test the model [29, 30].
Features univariately associated to response (pCR vs no pCR) were selected using WMW tests or t test, depending on the normality of data distribution, previously assessed using Shapiro–Wilk test [16, 22].
These tests were separately applied for the two cohorts of patients, and correction for multiple comparisons was performed by using the Benjamini–Hochberg method [18].
The correlation between significant features was evaluated in terms of Pearson Correlation Coefficient (PCC) [30].
The significant features were sorted in function of the p-value obtained at the univariate analysis. The feature showing the lowest p-value in the 1.5 T dataset was combined with the most significant one obtained at 3 T, and their combination was considered as starting point of the multivariate analysis.
Multiple logistic regression models were created adding as third feature one selected among those resulting significant at least in one of the two datasets.
The models were trained on the whole dataset, obtained merging 1.5 T and 3 T patients, and evaluated in terms of AUC and Aikake Information Criteria (AIC) [31]. The robustness of the predictive model was then evaluated using a tenfold cross-validation analysis with 300 repetitions as cross-validation method.
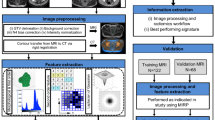
Finally, the model was also applied to the two separate datasets to evaluate its performance when used on MR images acquired at the same magnetic field intensity. In Fig. 1, the adopted workflow for feature selection is graphically depicted.
Schematic representation of the workflow adopted for data analysis: feature A was the most significant feature at 1.5 T; feature B was the most significant feature at 3 T; feature C was the feature that combined with feature A and B allowed to generate the predictive model with the highest AUC value
Results
The clinical characteristics of the patients enrolled in the study are reported in Table 1. The clinical tumour staging at the time of diagnosis was reported according to the TNM AJCC-UICC classification [32].
No statistically significant difference was observed between the two cohorts of patients. A total of 30 patients showed pCR (TRG = 1) in institution A, with a pCR rate equal to 22%. A similar pCR rate was obtained in institution B (25%) where 15 of 59 cases were complete responders.
Table 2 reports the five features showing the highest significance at the univariate analysis in the cohort of the patients acquired using a 1.5 T MR scanner.
Table 3 reports the same values obtained for patients acquired with 3 T MR scanner.
The features and the relative coefficients characterising the statistical model able to predict pCR are reported in Table 4.
The first two features selected in the model were the maximum FD, calculated on the pixel clusters individuated using the IB filter with 0% as lower level and 50% as upper level (IB: 0–50), and the energy calculated on the raw image.
The grey-level non-uniformity (GLNU) calculated on the run length matrix (GLNU_RLM) for the same pixel cluster chosen for maximum FD (IB: 0–50) was chosen as third feature, as it ensures the predictive model with the highest AUC.
The AIC value of the developed model was equal to 192.23, that was the lowest value obtained using 3 features. No additional features were added to the model, as the addition of a fourth feature resulted in increasing the AIC.
Figure 2 reports the ROC curves of the elaborated model. The AUC of the model tested on the whole dataset after the cross-validation was 0.72. The resulting AUC for the 1.5 T cohort was 0.70, while 0.83 for 3 T.
Discussion
In this study, a MR radiomics prediction model for pCR after nCRT in LARC was developed. The pCR rates observed in the two centres (22% in Institution A and 25% in Institution B) were consistent with the clinical data reported in the literature [7, 33].
To our knowledge, this predictive model is the first model able to show promising performances in pCR prediction mixing 1.5 T and 3 T MR images.
This study demonstrates that by using appropriate feature selection methods, it is possible to elaborate predictive models able to overcome the variability due to different magnetic field intensities.
The small p-values in Table 4 demonstrate that all the parameters in the model have discriminating powers. This indicates that with the strategy for feature selection and model elaboration presented in this study it is possible to create predictive models.
Different MR-based prediction models for response to nCRT for LARC patients have been published using images acquired at fixed magnetic field intensity [14, 16, 22, 34]. Intven et al. analysed 3 T MR images, combining features coming from different MR sequences (T2 weighted, diffusion weighted and dynamic contrast enhanced images) and different acquisition timing (before and after CRT) to predict pCR [35]. The authors obtained a very high accuracy (AUC = 0.93 analysing the difference on the ADC maps) but the analysis was done in a relative limited cohort of patients (55 cases) without external validation.
Dinapoli et al. trained a statistical model on an internal cohort of 162 patients and then validated the model on an external dataset of 49 patients from two different institutions. This model, based on the image intensities of T2 weighted MR images acquired before CRT with using 1.5 T MR scanners belonging to different vendors, reports AUC values of 0.73 for the internal and 0.75 for the external dataset [16]. Cusumano et al. added new features coming from fractal analysis to the aforementioned model, aiming to better describe the tumour heterogeneity in the case of LARC patients. The obtained predictive model integrating such fractal features reached AUC of 0.77 ± 0.07 in the internal dataset and 0.79 ± 0.09 in the external one [22]. The AUC of 0.72 obtained in this study combining 3 T and 1.5 T images was comparable to those found in the 1.5 T studies.
The univariate analysis carried out in this work has highlighted the superiority of the 3 T MR images in providing more predictive features respect to 1.5 T MR ones. The most significant feature at 1.5 T shows a p-value equal to 10−4 (maximum fractal dimension for pixels with intensity lower than 50%), while at 3 T p-value is 10−7 (energy without applying any image filter). The higher prognostic potentiality of 3 T MR images can be ascribable to the tighter spatial resolution of the images and the higher signal to noise ratio.
One of the limitations of this study is represented by the absence of an independent external dataset composed by MR images acquired at 1.5 T and 3 T in third institution. In absence of these data, the robustness of the model here proposed has been confirmed by the results obtained applying a cross-validation method using a high number of folders and repetitions. A perspective validation of the model proposed will be carried out in the next future, including also data from different centres. However, the results of this study demonstrate that most of the technical aspects that today limit the radiomics diffusion, such as the variability of MR scanners in field strengths and manufacturers, can be overcome by means of dedicated strategy of data analysis.
Conclusions
In conclusion, a MR radiomics prediction model for pCR after neoadjuvant therapy in locally advanced rectal cancer was developed: the model showed good performance, even when data from patients scanned on 1.5 T and 3 T were merged. This demonstrates that the magnetic field intensity variability can be overcome by means of selecting appropriate images features.
The possibility to overcome the inter-scanner variability and to predict pCR before CRT treatment opens innovative scenarios in cancer care, adding new evidence towards fully personalised approaches and treatment tailoring based on patient-specific tumour heterogeneity description.
Abbreviations
- AIC:
-
Aikake information criteria
- AUC:
-
Area under curve
- GLNU:
-
Grey-level non-uniformity
- IB:
-
Intensity based
- LARC:
-
Locally advanced rectal cancer
- LE:
-
Local excision
- LOG:
-
Laplacian of Gaussian
- MRI:
-
Magnetic resonance imaging
- nCRT:
-
Neoadjuvant chemo-radiotherapy
- pCR:
-
Pathological complete response
- ROI:
-
Region of interest
- T2-w:
-
T2-weighted
- TME:
-
Total mesorectal excision
- TRG:
-
Tumour regression grade
- W&W:
-
Watch and wait
- WMW:
-
Wilcoxon–Mann–Whitney
References
Berardi R, Maccaroni E, Onofri A et al (2014) Locally advanced rectal cancer: the importance of a multidisciplinary approach. World J Gastroenterol 20:17279–17287. https://doi.org/10.3748/wjg.v20.i46.17279
Tamas K, Walenkamp AME, de Vries EGE et al (2015) Rectal and colon cancer: not just a different anatomic site. Cancer Treat Rev 41:671–679. https://doi.org/10.1016/j.ctrv.2015.06.007
Belluco C, De Paoli A, Canzonieri V et al (2011) Long-term outcome of patients with complete pathologic response after neoadjuvant chemoradiation for cT3 rectal cancer: implications for local excision surgical strategies. Ann Surg Oncol 18:3686–3693. https://doi.org/10.1245/s10434-011-1822-0
Maas M, Nelemans PJ, Valentini V et al (2010) Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol 11:835–844. https://doi.org/10.1016/S1470-2045(10)70172-8
Martin ST, Heneghan HM, Winter DC (2012) Systematic review and meta-analysis of outcomes following pathological complete response to neoadjuvant chemoradiotherapy for rectal cancer. Br J Surg 99:918–928. https://doi.org/10.1002/bjs.8702
Ortholan C, Romestaing P, Chapet O, Gerard JP (2012) Correlation in rectal cancer between clinical tumor response after neoadjuvant radiotherapy and sphincter or organ preservation: 10-year results of the Lyon R 96-02 randomized trial. Int J Radiat Oncol Biol Phys 83:e165–e171. https://doi.org/10.1016/j.ijrobp.2011.12.002
Valentini V, van Stiphout RGPM, Lammering G et al (2015) Selection of appropriate end-points (pCR vs 2yDFS) for tailoring treatments with prediction models in locally advanced rectal cancer. Radiother Oncol 114:302–309. https://doi.org/10.1016/j.radonc.2015.02.001
Valentini V, van Stiphout RGPM, Lammering G et al (2011) Nomograms for predicting local recurrence, distant metastases, and overall survival for patients with locally advanced rectal cancer on the basis of European randomized clinical trials. J Clin Oncol 29:3163–3172. https://doi.org/10.1200/JCO.2010.33.1595
Dattani M, Heald RJ, Goussous G et al (2018) Oncological and survival outcomes in watch and wait patients with a clinical complete response after neoadjuvant chemoradiotherapy for rectal cancer: a systematic review and pooled analysis. Ann Surg. https://doi.org/10.1097/SLA.0000000000002761
Renehan AG, Malcomson L, Emsley R et al (2016) Watch-and-wait approach versus surgical resection after chemoradiotherapy for patients with rectal cancer (the OnCoRe project): a propensity-score matched cohort analysis. Lancet Oncol 17:174–183. https://doi.org/10.1016/S1470-2045(15)00467-2
Habr-Gama A, Perez RO, Nadalin W et al (2004) Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy: long-term results. Ann Surg 240:711–717 (discussion 717–718)
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577. https://doi.org/10.1148/radiol.2015151169
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446. https://doi.org/10.1016/j.ejca.2011.11.036
Hötker AM, Tarlinton L, Mazaheri Y et al (2016) Multiparametric MRI in the assessment of response of rectal cancer to neoadjuvant chemoradiotherapy: a comparison of morphological, volumetric and functional MRI parameters. Eur Radiol 26:4303–4312. https://doi.org/10.1007/s00330-016-4283-9
Patel UB, Brown G, Rutten H et al (2012) Comparison of magnetic resonance imaging and histopathological response to chemoradiotherapy in locally advanced rectal cancer. Ann Surg Oncol 19:2842–2852. https://doi.org/10.1245/s10434-012-2309-3
Dinapoli N, Barbaro B, Gatta R et al (2018) Magnetic resonance, vendor-independent, intensity histogram analysis predicting pathologic complete response after radiochemotherapy of rectal cancer. Int J Radiat Oncol Biol Phys. https://doi.org/10.1016/j.ijrobp.2018.04.065
Nie K, Shi L, Chen Q et al (2016) Rectal cancer: assessment of neoadjuvant chemoradiation outcome based on radiomics of multiparametric MRI. Clin Cancer Res 22:5256–5264. https://doi.org/10.1158/1078-0432.CCR-15-2997
Horvat N, Veeraraghavan H, Khan M et al (2018) MR imaging of rectal cancer: radiomics analysis to assess treatment response after neoadjuvant therapy. Radiology 287:833–843. https://doi.org/10.1148/radiol.2018172300
Nardone V, Reginelli A, Scala F et al (2019) Magnetic-resonance-imaging texture analysis predicts early progression in rectal cancer patients undergoing neoadjuvant chemoradiation. Gastroenterol Res Pract 2019:8505798. https://doi.org/10.1155/2019/8505798
Bergeest J-P, Jäger F (2008) A comparison of five methods for signal intensity standardization in MRI. In: Tolxdorff T, Braun J, Deserno TM et al (eds) Bildverarbeitung für die Medizin 2008. Springer, Berlin, pp 36–40
Dinapoli N, Casà C, Barbaro B (2016) Radiomics for rectal cancer. Transl Cancer Res 5:1–8
Cusumano D, Dinapoli N, Boldrini L et al (2018) Fractal-based radiomic approach to predict complete pathological response after chemo-radiotherapy in rectal cancer. Radiol Med 123:286–295. https://doi.org/10.1007/s11547-017-0838-3
Kumar V, Gu Y, Basu S et al (2012) Radiomics: the process and the challenges. Magn Reson Imaging 30:1234–1248. https://doi.org/10.1016/j.mri.2012.06.010
Liu Z, Zhang X-Y, Shi Y-J et al (2017) Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Clin Cancer Res 23:7253–7262. https://doi.org/10.1158/1078-0432.CCR-17-1038
Vecchio FM, Valentini V, Minsky BD et al (2005) The relationship of pathologic tumor regression grade (TRG) and outcomes after preoperative therapy in rectal cancer. Int J Radiat Oncol Biol Phys 62:752–760. https://doi.org/10.1016/j.ijrobp.2004.11.017
Grégoire V, Mackie TR (2011) State of the art on dose prescription, reporting and recording in intensity-modulated radiation therapy (ICRU report No. 83). Cancer Radiother 15:555–559. https://doi.org/10.1016/j.canrad.2011.04.003
Gatta R, Vallati M, Dinapoli N et al (2018) Towards a modular decision support system for radiomics: a case study on rectal cancer. Artif Intell Med. https://doi.org/10.1016/j.artmed.2018.09.003
Dinapoli N, Alitto AR, Vallati M et al (2015) Moddicom: a complete and easily accessible library for prognostic evaluations relying on image features. Conf Proc IEEE Eng Med Biol Soc 2015:771–774. https://doi.org/10.1109/EMBC.2015.7318476
Parmar C, Grossmann P, Bussink J et al (2015) Machine learning methods for quantitative radiomic biomarkers. Sci Rep 5:13087. https://doi.org/10.1038/srep13087
Taylor J (1997) An introduction to error analysis: the study of uncertainties in physical measurements, vol II. University Science Books, Sausalito
International Commissioning on Radiation Units and Measurements (2008) Receiver operating characteristic (ROC) analysis in medical imaging. ICRU Report 79
Brierley JD, Gospodarowicz MK, Wittekind C (2017) TNM classification of malignant tumours. Wiley, New York
Habr-Gama A, Gama-Rodrigues J, São Julião GP et al (2014) Local recurrence after complete clinical response and watch and wait in rectal cancer after neoadjuvant chemoradiation: impact of salvage therapy on local disease control. Int J Radiat Oncol Biol Phys 88:822–828. https://doi.org/10.1016/j.ijrobp.2013.12.012
Boldrini L, Cusumano D, Chiloiro G et al (2018) Delta radiomics for rectal cancer response prediction with hybrid 0.35 T magnetic resonance guided radiotherapy (MRgRT) : a hypothesis generating study for an innovative personalized medicine approach. La Radiologia Medica
Intven M, Monninkhof EM, Reerink O, Philippens MEP (2015) Combined T2w volumetry, DW-MRI and DCE-MRI for response assessment after neo-adjuvant chemoradiation in locally advanced rectal cancer. Acta Oncol 54:1729–1736. https://doi.org/10.3109/0284186X.2015.1037010
Acknowledgments
None.
Funding
Open access funding provided by Università Cattolica del Sacro Cuore within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors are not aware of any actual or potential conflict of interest in the field.
Research involving human participants
All procedures performed in this study were in accordance with the ethical standards of the institutional Italian committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study. This retrospective study has involved fully anonymized data according with the RGDP n. 2016/679.
Ethical approval
All the data were previously anonymised and belonged to patients who had consented to the use of their anonymized data for research purposes. A notification of the study was submitted and approved by the ethical committee of the institutions, as required by the national laws.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cusumano, D., Meijer, G., Lenkowicz, J. et al. A field strength independent MR radiomics model to predict pathological complete response in locally advanced rectal cancer. Radiol med 126, 421–429 (2021). https://doi.org/10.1007/s11547-020-01266-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11547-020-01266-z