Abstract
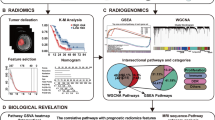
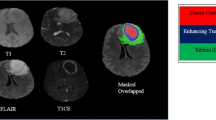
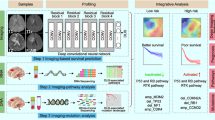
Glioblastoma multiforme (GBM) is one of the deadliest tumours. This study aimed to construct radiogenomic prognostic models of glioblastoma overall survival (OS) based on magnetic resonance imaging (MRI) Gd-T1WI images and deoxyribonucleic acid (DNA) methylation-seq and to understand the related biological pathways. The ResNet3D-18 model was used to extract radiomic features, and Lasso-Cox regression analysis was utilized to establish the prognostic models. A nomogram was constructed by combining the radiogenomic features and clinicopathological variables. The DeLong test was performed to compare the area under the curve (AUC) of the models. We screened differentially expressed genes (DEGs) with original ribonucleic acid (RNA)-seq in risk stratification and used Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) annotations for functional enrichment analysis. For the 1-year OS models, the AUCs of the radiogenomic set, methylation set and deep learning set in the training cohort were 0.864, 0.804 and 0.787, and those in the validation cohort were 0.835, 0.768 and 0.651, respectively. The AUCs of the 0.5-, 1- and 2-year nomograms in the training cohort were 0.943, 0.861 and 0.871, and those in the validation cohort were 0.864, 0.885 and 0.805, respectively. A total of 245 DEGs were screened; functional enrichment analysis showed that these DEGs were associated with cell immunity. The survival risk-stratifying radiogenomic models for glioblastoma OS had high predictability and were associated with biological pathways related to cell immunity.
Graphical Abstract








Similar content being viewed by others
References
Beig N, Bera K, Prasanna P, Antunes J, Correa R, Singh S et al (2020) Radiogenomic-based survival risk stratification of tumor habitat on Gd-T1w MRI is associated with biological processes in glioblastoma. Clin Cancer Res 26(8):1866–1876. https://doi.org/10.1158/1078-0432.CCR-19-2556
Zhang X, Lu H, Tian Q, Feng N, Yin L, Xu X et al (2019) A radiomics nomogram based on multiparametric MRI might stratify glioblastoma patients according to survival. Eur Radiol 29(10):5528–5538. https://doi.org/10.1007/s00330-019-06069-z
X. Jia, Y. Zhai, D. Song, Y. Wang, S. Wei, F. Yang, et al. (2022) A multiparametric MRI-based radiomics nomogram for preoperative prediction of survival stratification in glioblastoma patients with standard treatment. Frontiers in Oncology 12. https://doi.org/10.3389/fonc.2022.758622
J. Lao, Y. Chen, Z.-C. Li, Q. Li, J. Zhang, J. Liu, et al. (2017) A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Scientific Reports 7(1). https://doi.org/10.1038/s41598-017-10649-8
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D et al (2021) The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol 23(8):1231–1251. https://doi.org/10.1093/neuonc/noab106
Suelves M, Carrio E, Nunez-Alvarez Y, Peinado MA (2016) DNA methylation dynamics in cellular commitment and differentiation. Brief Funct Genomics 15(6):443–453. https://doi.org/10.1093/bfgp/elw017
Lu C, Wei Y, Wang X, Zhang Z, Yin J, Li W et al (2020) DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Mol Cancer 19(1):28. https://doi.org/10.1186/s12943-020-1137-5
Ji J, Zhao L, Zhao X, Li Q, An Y, Li L et al (2020) Genomewide DNA methylation regulation analysis of long noncoding RNAs in glioblastoma. Int J Mol Med 46(1):224–238. https://doi.org/10.3892/ijmm.2020.4579
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P et al (2013) The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26(6):1045–1057. https://doi.org/10.1007/s10278-013-9622-7
Tomczak K, Czerwinska P, Wiznerowicz M (2015) The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 19(1A):A68-77. https://doi.org/10.5114/wo.2014.47136
Zhang W, Peng J, Zhao S, Wu W, Yang J, Ye J et al (2022) Deep learning combined with radiomics for the classification of enlarged cervical lymph nodes. J Cancer Res Clin Oncol 148(10):2773–2780. https://doi.org/10.1007/s00432-022-04047-5
Ellingson BM, Abrey LE, Nelson SJ, Kaufmann TJ, Garcia J, Chinot O et al (2018) Validation of postoperative residual contrast-enhancing tumor volume as an independent prognostic factor for overall survival in newly diagnosed glioblastoma. Neuro Oncol 20(9):1240–1250. https://doi.org/10.1093/neuonc/noy053
Li G, Li L, Li Y, Qian Z, Wu F, He Y et al (2022) An MRI radiomics approach to predict survival and tumour-infiltrating macrophages in gliomas. Brain 145(3):1151–1161. https://doi.org/10.1093/brain/awab340
Koch A, Joosten SC, Feng Z, de Ruijter TC, Draht MX, Melotte V et al (2018) Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol 15(7):459–466. https://doi.org/10.1038/s41571-018-0004-4
Reis AH, Vargas FR, Lemos B (2016) Biomarkers of genome instability and cancer epigenetics. Tumour Biol 37(10):13029–13038. https://doi.org/10.1007/s13277-016-5278-5
Adeberg S, Knoll M, Koelsche C, Bernhardt D, Schrimpf D, Sahm F et al (2022) DNA-methylome-assisted classification of patients with poor prognostic subventricular zone associated IDH-wildtype glioblastoma. Acta Neuropathol 144(1):129–142. https://doi.org/10.1007/s00401-022-02443-2
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G et al (2017) m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep 18(11):2622–2634. https://doi.org/10.1016/j.celrep.2017.02.059
Huang W, Weng W, Wu B, Ye T, Lin Z, Zhang Z et al (2021) Development and validation of the trans-omics model for pancreatic adenocarcinoma. Epigenomics 13(1):15–30. https://doi.org/10.2217/epi-2020-0184
Chang S, Yim S, Park H (2019) The cancer driver genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism. Exp Mol Med 51(6):1–17. https://doi.org/10.1038/s12276-019-0230-6
L.B. Wang, A. Karpova, M.A. Gritsenko, J.E. Kyle, S. Cao, Y. Li, et al. (2021) Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell 39(4):509–528 e520. https://doi.org/10.1016/j.ccell.2021.01.006
Sugita Y, Nakamura Y, Yamamoto M, Ogasawara S, Ohshima K, Shigemori M (2008) Expression of KIAA 0864 protein in neuroepithelial tumors: an analysis based on the presence of monoclonal antibody HFB-16. J Neurooncol 89(2):151–158. https://doi.org/10.1007/s11060-008-9610-9
She S, Bian S, Huo R, Chen K, Huang Z, Zhang J et al (2016) Degradable organically-derivatized polyoxometalate with enhanced activity against glioblastoma cell line. Sci Rep 6:33529. https://doi.org/10.1038/srep33529
Dao LM, Machule ML, Bacher P, Hoffmann J, Ly LT, Wegner F et al (2021) Decreased inflammatory cytokine production of antigen-specific CD4(+) T cells in NMDA receptor encephalitis. J Neurol 268(6):2123–2131. https://doi.org/10.1007/s00415-020-10371-y
Niibori-Nambu A, Midorikawa U, Mizuguchi S, Hide T, Nagai M, Komohara Y et al (2013) Glioma initiating cells form a differentiation niche via the induction of extracellular matrices and integrin alphaV. PLoS ONE 8(5):e59558. https://doi.org/10.1371/journal.pone.0059558
Liu X, Li Y, Qian Z, Sun Z, Xu K, Wang K et al (2018) A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. Neuroimage Clin 20:1070–1077. https://doi.org/10.1016/j.nicl.2018.10.014
W. Meng, J.D. Palmer, M. Siedow, S.J. Haque, A. Chakravarti (2022) Overcoming radiation resistance in gliomas by targeting metabolism and DNA repair pathwayS. Int J Mol Sci 23(4). https://doi.org/10.3390/ijms23042246
David BA, Kubes P (2019) Exploring the complex role of chemokines and chemoattractants in vivo on leukocyte dynamics. Immunol Rev 289(1):9–30. https://doi.org/10.1111/imr.12757
Li M, Chen H, Yin P, Song J, Jiang F, Tang Z et al (2021) Identification and clinical validation of key extracellular proteins as the potential biomarkers in relapsing-remitting multiple sclerosis. Front Immunol 12:753929. https://doi.org/10.3389/fimmu.2021.753929
J.Y. Cao, Q. Guo, G.F. Guan, C. Zhu, C.Y. Zou, L.Y. Zhang, et al. (2020) Elevated lymphocyte specific protein 1 expression is involved in the regulation of leukocyte migration and immunosuppressive microenvironment in glioblastoma. Aging (Albany NY) 12(2):1656–1684. https://doi.org/10.18632/aging.102706
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical statement
All data involved in this study were downloaded from TCGA, and data collection and application were conducted in accordance with TCGA’s publication guidelines and data access policies, with additional approval from the local ethics committee.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, W., Yan, Z., Peng, J. et al. Magnetic resonance imaging and deoxyribonucleic acid methylation–based radiogenomic models for survival risk stratification of glioblastoma. Med Biol Eng Comput 62, 853–864 (2024). https://doi.org/10.1007/s11517-023-02971-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02971-3