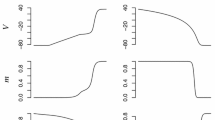
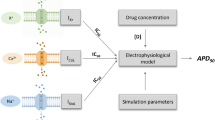
Abstract
Customization of cardiac action potential models has become increasingly important with the recognition of patient-specific models and virtual patient cohorts as valuable predictive tools. Nevertheless, developing customized models by fitting parameters to data poses technical and methodological challenges: despite noise and variability associated with real-world datasets, traditional optimization methods produce a single “best-fit” set of parameter values. Bayesian estimation methods seek distributions of parameter values given the data by obtaining samples from the target distribution, but in practice widely known Bayesian algorithms like Markov chain Monte Carlo tend to be computationally inefficient and scale poorly with the dimensionality of parameter space. In this paper, we consider two computationally efficient Bayesian approaches: the Hamiltonian Monte Carlo (HMC) algorithm and the approximate Bayesian computation sequential Monte Carlo (ABC-SMC) algorithm. We find that both methods successfully identify distributions of model parameters for two cardiac action potential models using model-derived synthetic data and an experimental dataset from a zebrafish heart. Although both methods appear to converge to the same distribution family and are computationally efficient, HMC generally finds narrower marginal distributions, while ABC-SMC is less sensitive to the algorithmic settings including the prior distribution.
Graphical abstract












Similar content being viewed by others
Availability of data and material
The data used for this study are available upon request.
Code availability
The code used for this study is available upon request.
References
Fenton FH, Cherry EM (2008) Models of cardiac cell. Scholarpedia 3(8):1868
Groenendaal W, Ortega FA, Kherlopian AR, Zygmunt AC, Krogh-Madsen T, Christini DJ (2015) Cell-specific cardiac electrophysiology models. PLoS Comput Biol 11(4):1004242. https://doi.org/10.1371/journal.pcbi.1004242
Boyle PM, Zghaib T, Zahid S, Ali RL, Deng D, Franceschi WH, Hakim JB, Murphy MJ, Prakosa A, Zimmerman SL, Ashikaga H, Marine JE, Kolandaivelu A, Nazarian S, Spragg DD, Calkins H, Trayanova NA (2019) Computationally guided personalized targeted ablation of persistent atrial fibrillation. Nat Biomed Eng 3(11):870–879
Niederer SA, Aboelkassem Y, Cantwell CD, Corrado C, Coveney S, Cherry EM, Delhaas T, Fenton FH, Panfilov AV, Pathmanathan P, Plank G, Riabiz M, Roney CH, dos Santos RW, Wang L (2020) Creation and application of virtual patient cohorts of heart models. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 378(2173):20190558. https://doi.org/10.1098/rsta.2019.0558
Dokos S, Lovell NH (2004) Parameter estimation in cardiac ionic models. Prog Biophys Mol Biol 85(2):407–431. https://doi.org/10.1016/j.pbiomolbio.2004.02.002
Bueno-Orovio A, Cherry EM, Fenton FH (2008) Minimal model for human ventricular action potentials in tissue. J Theor Biol 253(3):544–560. https://doi.org/10.1016/j.jtbi.2008.03.029
Syed Z, Vigmond E, Nattel S, Leon LJ (2005) Atrial cell action potential parameter fitting using genetic algorithms. Med Biol Eng Compu 43(5):561–571
Bot CT, Kherlopian AR, Ortega FA, Christini DJ, Krogh-Madsen T (2012) Rapid genetic algorithm optimization of a mouse computational model: benefits for anthropomorphization of neonatal mouse cardiomyocytes. Front Physiol 3:421. https://doi.org/10.3389/fphys.2012.00421
Cairns DI, Fenton FH, Cherry EM (2017) Efficient parameterization of cardiac action potential models using a genetic algorithm. Chaos 27(9):093922. https://doi.org/10.1063/1.5000354
Loewe A, Wilhelms M, Schmid J, Krause MJ, Fischer F, Thomas D, Scholz EP, Dössel O, Seemann G (2015) Parameter estimation of ion current formulations requires hybrid optimization approach to be both accurate and reliable. Front Bioeng Biotechnol 3:209. https://doi.org/10.3389/fbioe.2015.00209
Coveney S, Clayton RH (2018) Fitting two human atrial cell models to experimental data using Bayesian history matching. Prog Biophys Mol Biol 139:43–58. https://doi.org/10.1016/j.pbiomolbio.2018.08.001
Zaman MS, Dhamala J, Bajracharya P, Sapp JL, Horácek BM, Wu KC, Trayanova NA, Wang L (2021) Fast posterior estimation of cardiac electrophysiological model parameters via Bayesian active learning. Frontiers in Physiology 12: 740306. https://doi.org/10.3389/fphys.2021.740306.Accessed 2022-04-15
Siekmann I, Wagner LE, Yule D, Fox C, Bryant D, Crampin EJ, Sneyd J (2011) MCMC estimation of Markov models for ion channels. Biophys J 100(8):1919–1929. https://doi.org/10.1016/j.bpj.2011.02.059
Pathmanathan P, Shotwell MS, Gavaghan DJ, Cordeiro JM, Gray RA (2015) Uncertainty quantification of fast sodium current steady-state inactivation for multi-scale models of cardiac electrophysiology. Prog Biophys Mol Biol 117(1):4–18. https://doi.org/10.1016/j.pbiomolbio.2015.01.008
Daly AC, Gavaghan DJ, Holmes C, Cooper J (2015) Hodgkin-Huxley revisited: reparametrization and identifiability analysis of the classic action potential model with approximate Bayesian methods. Royal Society Open Science 2(12):150499. https://doi.org/10.1098/rsos.150499
Neal, R.M.: MCMC using Hamiltonian dynamics. In: Handbook of Markov chain Monte Carlo. Chapman & Hall/CRC Handb. Mod. Stat. Methods, pp. 113–162 (2011)
Vernon I, Liu J, Goldstein M, Rowe J, Topping J, Lindsey K (2018) Bayesian uncertainty analysis for complex systems biology models: emulation, global parameter searches and evaluation of gene functions. BMC Syst Biol 12(1):1. https://doi.org/10.1186/s12918-017-0484-3
Del Moral P, Doucet A, Jasra A (2006) Sequential Monte Carlo samplers. Journal of the Royal Statistical Society. Series B (Statistical Methodology) 68(3): 411–436
Toni T, Welch D, Strelkowa N, Ipsen A, Stumpf MPH (2009) Approximate Bayesian computation scheme for parameter inference and model selection in dynamical systems. J R Soc Interface 6(31):187–202. https://doi.org/10.1098/rsif.2008.0172
Del Moral P, Doucet A, Jasra A (2012) An adaptive sequential Monte Carlo method for approximate Bayesian computation. Stat Comput 22(5):1009–1020. https://doi.org/10.1007/s11222-011-9271-y
O’Hara T, Virág L, Varró A, Rudy Y (2011) Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput Biol 7(5):1002061
Daly AC, Cooper J, Gavaghan DJ, Holmes C (2017) Comparing two sequential Monte Carlo samplers for exact and approximate Bayesian inference on biological models. J R Soc Interface 14(134):20170340. https://doi.org/10.1098/rsif.2017.0340
Duane S, Kennedy AD, Pendleton BJ, Roweth D (1987) Hybrid Monte Carlo. Phys Lett B 195(2):216–222. https://doi.org/10.1016/0370-2693(87)91197-X
Monnahan CC, Thorson JT, Branch TA (2017) Faster estimation of Bayesian models in ecology using Hamiltonian Monte Carlo. Methods Ecol Evol 8(3):339–348. https://doi.org/10.1111/2041-210X.12681
Margossian CC, Zhang Y, Gillespie WR (2021) Flexible and efficient Bayesian pharmacometrics modeling using Stan and Torsten, part I. arXiv:2109.10184 [stat]
Nieto Ramos A, Herndon CJ, Fenton FH, Cherry EM (2021) Quantifying distributions of parameters for cardiac action potential models using the Hamiltonian Monte Carlo method. Computing in Cardiology 48:9662836–196628364
Mitchell CC, Schaeffer DG (2003) A two-current model for the dynamics of cardiac membrane. Bull Math Biol 65(5):767–793. https://doi.org/10.1016/S0092-8240(03)00041-7
Fenton F, Karma A (1998) Vortex dynamics in three-dimensional continuous myocardium with fiber rotation: filament instability and fibrillation. Chaos 8(1):20–47. https://doi.org/10.1063/1.166311
Fenton FH, Cherry EM, Hastings HM, Evans SJ (2002) Multiple mechanisms of spiral wave breakup in a model of cardiac electrical activity. Chaos 12(3):852–892. https://doi.org/10.1063/1.1504242
Gamerman D, Lopes HF (2006) Markov chain Monte Carlo: stochastic simulation for Bayesian inference, Second Edition
Marin J-M, Pudlo P, Robert CP, Ryder RJ (2012) Approximate Bayesian computational methods. Stat Comput 22(6):1167–1180. https://doi.org/10.1007/s11222-011-9288-2
Betancourt M (2018) A conceptual introduction to Hamiltonian Monte Carlo. arXiv:1701.02434 [stat]
Hoffman MD, Gelman A (2014) The No-U-turn sampler: adaptively setting path lengths in Hamiltonian Monte Carlo. J Mach Learn Res 15(1):1593–1623
Carpenter B, Gelman A, Hoffman MD, Lee D, Goodrich B, Betancourt M, Brubaker M, Guo J, Li P, Riddell A (2017) Stan: a probabilistic programming language. J Stat Softw 76:1–32. https://doi.org/10.18637/jss.v076.i01
Stan Development Team (2022) Stan modeling language users guide and reference manual, version 2.29. //mc-stan.org/
Gelman A, Carlin JB, Stern HS, Dunson DB, Vehtari A, Rubin DB (2013) Bayesian data analysis, 3rd edition edn. Chapman and Hall/CRC
Vehtari A, Gelman A, Simpson D, Carpenter B, Bürkner P-C (2021) Rank-normalization, folding, and localization: an improved \(\widehat{R}\) for assessing convergence of MCMC. Bayesian Analysis 16(2). https://doi.org/10.1214/20-BA1221
Chicco D, Warrens MJ, Jurman G (2021) The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ Computer Science 7:623. https://doi.org/10.7717/peerj-cs.623
Shahi S, Marcotte CD, Herndon CJ, Fenton FH, Shiferaw Y, Cherry EM (2021) Long-time prediction of arrhythmic cardiac action potentials using recurrent neural networks and reservoir computing. Frontiers in physiology 12
Whittaker DG, Clerx M, Lei CL, Christini DJ, Mirams GR (2020) Calibration of ionic and cellular cardiac electrophysiology models. Wiley Interdisciplinary Reviews: Systems Biology and Medicine 12(4):1482
Johnstone RH, Chang ET, Bardenet R, De Boer TP, Gavaghan DJ, Pathmanathan P, Clayton RH, Mirams GR (2016) Uncertainty and variability in models of the cardiac action potential: can we build trustworthy models? J Mol Cell Cardiol 96:49–62
Britton OJ, Bueno-Orovio A, Van Ammel K, Lu HR, Towart R, Gallacher DJ, Rodriguez B (2013) Experimentally calibrated population of models predicts and explains intersubject variability in cardiac cellular electrophysiology. Proc Natl Acad Sci USA 110(23):2098–2105. https://doi.org/10.1073/pnas.1304382110
Csercsik D, Hangos KM, Szederkényi G (2012) Identifiability analysis and parameter estimation of a single Hodgkin-Huxley type voltage dependent ion channel under voltage step measurement conditions. Neurocomputing 77(1):178–188. https://doi.org/10.1016/j.neucom.2011.09.006
Shotwell MS, Gray RA (2016) Estimability analysis and optimal design in dynamic multi-scale models of cardiac electrophysiology. J Agric Biol Environ Stat 21(2):261–276. https://doi.org/10.1007/s13253-016-0244-7
Daly AC, Gavaghan D, Cooper J, Tavener S (2018) Inference-based assessment of parameter identifiability in nonlinear biological models. J R Soc Interface 15(144):20180318. https://doi.org/10.1098/rsif.2018.0318
Chang KC, Dutta S, Mirams GR, Beattie KA, Sheng J, Tran PN, Wu M, Wu WW, Colatsky T, Strauss DG, Li Z (2017) Uncertainty quantification reveals the importance of data variability and experimental design considerations for in silico proarrhythmia risk assessment. Front Physiol 8:917. https://doi.org/10.3389/fphys.2017.00917
Funding
This study was supported by NSF grants CNS-2028677 and CMMI-1762553 and by NIH grant 1R01HL143450.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Data collection and analysis were performed by Alejandro Nieto Ramos. The first draft of the manuscript was written by Alejandro Nieto Ramos and all authors contributed to and commented on subsequent versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nieto Ramos, A., Fenton, F.H. & Cherry, E.M. Bayesian inference for fitting cardiac models to experiments: estimating parameter distributions using Hamiltonian Monte Carlo and approximate Bayesian computation. Med Biol Eng Comput 61, 75–95 (2023). https://doi.org/10.1007/s11517-022-02685-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-022-02685-y