Abstract
BACKGROUND
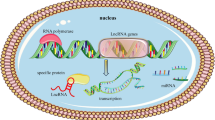
The majority of mammalian genomes have been found to be transcribed into non-coding RNAs. One category of non-coding RNAs is classified as long non-coding RNAs (lncRNAs) based on their transcript sizes larger than 200 nucleotides. Growing evidence has shown that lncRNAs are not junk transcripts and play regulatory roles in multiple aspects of biological processes. Dysregulation of lncRNA expression has also been linked to diseases, in particular cancer. Therefore, studies of lncRNAs have attracted significant interest in the field of medical research. Nuclear enriched abundant transcript 1 (NEAT1), a nuclear lncRNA, has recently emerged as a key regulator involved in various cellular processes, physiological responses, developmental processes, and disease development and progression.
OBJECTIVE
This review will summarize and discuss the most recent findings with regard to the roles of NEAT1 in the function of the nuclear paraspeckle, cellular pathways, and physiological responses and processes. Particularly, the most recently reported studies regarding the pathological roles of deregulated NEAT1 in cancer are highlighted in this review.
METHODS
We performed a systematic literature search using the Pubmed search engine. Studies published over the past 8 years (between January 2009 and August 2016) were the sources of literature review. The following keywords were used: “Nuclear enriched abundant transcript 1,” “NEAT1,” and “paraspeckles.”
RESULTS
The Pubmed search identified 34 articles related to the topic of the review. Among the identified literature, 13 articles report findings related to cellular functions of NEAT1 and eight articles are the investigations of physiological functions of NEAT1. The remaining 13 articles are studies of the roles of NEAT1 in cancers.
CONCLUSION
Recent advances in NEAT1 studies reveal the multifunctional roles of NEAT1 in various biological processes, which are beyond its role in nuclear paraspeckles. Recent studies also indicate that dysregulation of NEAT1 function contributes to the development and progression of various cancers. More investigations will be needed to address the detailed mechanisms regarding how NEAT1 executes its cellular and physiological functions and how NEAT1 dysregulation results in tumorigenesis, and to explore the potential of NEAT1 as a target in cancer diagnosis, prognosis and therapy.
Similar content being viewed by others
References
Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W, Leucci E, Lapouge G, Beck B, van den Oord J, Nakagawa S, Hirose T, Sablina A A, Lambrechts D, Aerts S, Blanpain C, Marine J C (2016). p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med, 22(8): 861–868
Arredouani M S, Lu B, Bhasin M, Eljanne M, Yue W, Mosquera J M, Bubley G J, Li V, Rubin M A, Libermann T A, Sanda M G (2009). Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res, 15(18): 5794–5802
Athanasiadis A, Rich A, Maas S (2004). Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol, 2 (12): e391
Birney E, Stamatoyannopoulos J A, Dutta A, Guigó R, Gingeras T R, Margulies E H, Weng Z, Snyder M, Dermitzakis E T, Thurman R E, KuehnMS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum J A, Andrews R M, Flicek P, Boyle P J, Cao H, Carter N P, Clelland G K, Davis S, Day N, Dhami P, Dillon S C, Dorschner M O, Fiegler H, Giresi P G, Goldy J, Hawrylycz M, Haydock A, Humbert R, James K D, Johnson B E, Johnson E M, Frum T T, Rosenzweig E R, Karnani N, Lee K, Lefebvre G C, Navas P A, Neri F, Parker S C, Sabo P J, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins F S, Dekker J, Lieb J D, Tullius T D, Crawford G E, Sunyaev S, Noble W S, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker I L, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch H A, Sekinger E A, Lagarde J, Abril J F, Shahab A, Flamm C, Fried C, Hackermüller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen J S, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas D J, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan K G, Sung WK, Ooi H S, Chiu K P, Foissac S, Alioto T, Brent M, Pachter L, Tress M L, Valencia A, Choo S W, Choo C Y, Ucla C, Manzano C, Wyss C, Cheung E, Clark T G, Brown J B, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen C N, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick J S, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers R M, Rogers J, Stadler P F, Lowe T M, Wei C L, Ruan Y, Struhl K, Gerstein M, Antonarakis S E, Fu Y, Green E D, Karaöz U, Siepel A, Taylor J, Liefer L A, Wetterstrand K A, Good P J, Feingold E A, Guyer M S, Cooper G M, Asimenos G, Dewey C N, Hou M, Nikolaev S, Montoya-Burgos J I, Löytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang N R, Holmes I, Mullikin J C, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent W J, Stone E A, Batzoglou S, Goldman N, Hardison R C, Haussler D, Miller W, Sidow A, Trinklein N D, Zhang Z D, Barrera L, Stuart R, King D C, Ameur A, Enroth S, Bieda M C, Kim J, Bhinge A A, Jiang N, Liu J, Yao F, Vega V B, Lee CW, Ng P, Shahab A, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley M J, Inman D, Singer M A, Richmond T A, Munn K J, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler J C, Couttet P, Bruce A W, Dovey O M, Ellis P D, Langford C F, Nix D A, Euskirchen G, Hartman S, Urban A E, Kraus P, Van Calcar S, Heintzman N, Kim T H, Wang K, Qu C, Hon G, Luna R, Glass C K, Rosenfeld M G, Aldred S F, Cooper S J, Halees A, Lin JM, Shulha H P, Zhang X, Xu M, Haidar J N, Yu Y, Ruan Y, Iyer V R, Green R D, Wadelius C, Farnham P J, Ren B, Harte R A, Hinrichs A S, Trumbower H, Clawson H, Hillman-Jackson J, Zweig A S, Smith K, Thakkapallayil A, Barber G, Kuhn R M, Karolchik D, Armengol L, Bird C P, de Bakker P I, Kern A D, Lopez-Bigas N, Martin J D, Stranger B E, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrímsdóttir I B, Huppert J, Zody M C, Abecasis G R, Estivill X, Bouffard G G, Guan X, Hansen N F, Idol J R, Maduro V V, Maskeri B, McDowell J C, Park M, Thomas P J, Young A C, Blakesley R W, Muzny D M, Sodergren E, Wheeler D A, Worley K C, Jiang H, Weinstock G M, Gibbs R A, Graves T, Fulton R, Mardis E R, Wilson R K, Clamp M, Cuff J, Gnerre S, Jaffe D B, Chang J L, Lindblad-Toh K, Lander E S, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong P J, and the ENCODE Project Consortium, the NISC Comparative Sequencing Program, the Baylor College of Medicine Human Genome Sequencing Center, the Washington University Genome Sequencing Center, the Broad Institute, the Children’s Hospital Oakland Research Institute (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature, 447(7146): 799–816
Bond C S, Fox A H (2009). Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol, 186(5): 637–644
Buckley N E, Mullan P B (2012). BRCA1–conductor of the breast stem cell orchestra: the role of BRCA1 in mammary gland development and identification of cell of origin of BRCA1 mutant breast cancer. Stem Cell Rev, 8(3): 982–993
Cardinale S, Cisterna B, Bonetti P, Aringhieri C, Biggiogera M, Barabino S M (2007). Subnuclear localization and dynamics of the Pre-mRNA 3' end processing factor mammalian cleavage factor I 68-kDa subunit. Mol Biol Cell, 18(4): 1282–1292
Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk A M, Chiu K P, Choudhary V, Christoffels A, Clutterbuck D R, Crowe M L, Dalla E, Dalrymple B P, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher C F, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras T R, Gojobori T, Green R E, Gustincich S, Harbers M, Hayashi Y, Hensch T K, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan S P, Kruger A, Kummerfeld S K, Kurochkin IV, Lareau L F, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid J F, Ring B Z, Ringwald M, Rost B, Ruan Y, Salzberg S L, Sandelin A, Schneider C, Schönbach C, Sekiguchi K, Semple C A, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan S L, Tang S, Taylor MS, Tegner J, Teichmann S A, Ueda H R, Van Nimwegen E, Verardo R, Wei C L, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond S M, Teasdale R D, Liu E T, Brusic V, Quackenbush J, Wahlestedt C, Mattick J S, Hume D A, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y; FANTOM Consortium.; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) (2005). The transcriptional landscape of the mammalian genome. Science, 309(5740): 1559–1563
Chai Y, Liu J, Zhang Z, Liu L (2016). HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med, 5(7): 1588–1598
Chakravarty D, Sboner A, Nair S S, Giannopoulou E, Li R, Hennig S, Mosquera J M, Pauwels J, Park K, Kossai M, MacDonald T Y, Fontugne J, Erho N, Vergara I A, Ghadessi M, Davicioni E, Jenkins R B, Palanisamy N, Chen Z, Nakagawa S, Hirose T, Bander N H, Beltran H, Fox A H, Elemento O, Rubin M A (2014). The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun, 5: 5383
Chen L L, Carmichael G G (2009). Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell, 35(4): 467–478
Chen L L, Carmichael G G (2010). Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol, 22(3): 357–364
Chen L L, De Cerbo J N, Carmichael G G (2008). Alu element-mediated gene silencing. EMBO J, 27(12): 1694–1705
Chen X, Kong J, Ma Z, Gao S, Feng X (2015). Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am J Cancer Res, 5(9): 2808–2815
Choudhry H, Albukhari A, Morotti M, Haider S, Moralli D, Smythies J, Schödel J, Green C M, Camps C, Buffa F, Ratcliffe P, Ragoussis J, Harris A L, Mole D R (2015). Tumor hypoxia induces nuclear paraspeckle formation through HIF-2a dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene, 34 (34): 4546
Chujo T, Yamazaki T, Hirose T (2016). Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim Biophys Acta, 1859(1): 139–146
Clemson C M, Hutchinson J N, Sara S A, Ensminger A W, Fox A H, Chess A, Lawrence J B (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell, 33(6): 717–726
Cooper D R, Carter G, Li P, Pate R, Watson J E, Patel N A (2014). Long Non-Coding RNA NEAT1 Associates with SRp40 to Temporally Regulate PPARg2 Splicing during Adipogenesis in 3T3-L1 Cells. Genes (Basel), 5(4): 1050–1063
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles D G, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown J B, Lipovich L, Gonzalez J M, Thomas M, Davis C A, Shiekhattar R, Gingeras T R, Hubbard T J, Notredame C, Harrow J, Guigó R (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res, 22(9): 1775–1789
Doria M, Neri F, Gallo A, Farace M G, Michienzi A (2009). Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res, 37(17): 5848–5858
Eidem T M, Kugel J F, Goodrich J A (2016). Noncoding RNAs: regulators of the mammalian transcription machinery. J Mol Biol, 428(12): 2652–2659
Foulkes W D (2004). BRCA1 functions as a breast stem cell regulator. J Med Genet, 41(1): 1–5
Fox A H, Bond C S, Lamond A I (2005). P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell, 16(11): 5304–5315
Fox A H, Lam Y W, Leung A K, Lyon C E, Andersen J, Mann M, Lamond A I (2002). Paraspeckles: a novel nuclear domain. Curr Biol, 12(1): 13–25
Fu J W, Kong Y, Sun X (2016). Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol, 142(7): 1571–1579
Gernapudi R, Wolfson B, Zhang Y, Yao Y, Yang P, Asahara H, Zhou Q (2015). MicroRNA 140 promotes expression of long noncoding RNA NEAT1 in adipogenesis. Mol Cell Biol, 36(1): 30–38
Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong M, Dang Y, Feng Z, Chen G (2015). Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol, 8(5): 5395–5402
Guru S C, Agarwal S K, Manickam P, Olufemi S E, Crabtree J S, Weisemann JM, Kester MB, Kim Y S, Wang Y, Emmert-BuckMR, Liotta L A, Spiegel AM, Boguski MS, Roe B A, Collins F S, Marx S J, Burns L, Chandrasekharappa S C (1997). A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res, 7(7): 725–735
He C, Jiang B, Ma J, Li Q (2016). Aberrant NEAT1 expression is associated with clinical outcome in high grade glioma patients. APMIS, 124(3): 169–174
Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Bénard M, Fox A H, Pierron G (2014). NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell, 25(1): 169–183
Hutchinson J N, Ensminger AW, Clemson CM, Lynch C R, Lawrence J B, Chess A (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics, 8 (1): 39
Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, Kai C, Yada T, Suzuki Y, Yamada T, Ozawa T, Kaneki K, Inoue T, Kobayashi M, Kodama T, Wada Y, Sekimizu K, Akimitsu N (2014). Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell, 53(3): 393–406
International Human Genome Sequencing Consortium (2004). Finishing the euchromatic sequence of the human genome. Nature, 431(7011): 931–945
Iyer MK, Niknafs Y S, Malik R, Singhal U, Sahu A, Hosono Y, Barrette T R, Prensner J R, Evans J R, Zhao S, Poliakov A, Cao X, Dhanasekaran S M, Wu Y M, Robinson D R, Beer D G, Feng F Y, Iyer H K, Chinnaiyan A M (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat Genet, 47(3): 199–208
Jiang P, Wu X, Wang X, Huang W, Feng Q (2016). NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget, 7(28): 43337–43351
Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR (2007). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science, 316(5830): 1484–1488
Ke H, Zhao L, Feng X, Xu H, Zou L, Yang Q, Su X, Peng L, Jiao B (2016). NEAT1 is required for survival of breast cancer cells through FUS and miR-548. Gene Regul Syst Bio, 10 (Suppl 1): 11–17
Kellis M, Wold B, Snyder MP, Bernstein B E, Kundaje A, Marinov G K, Ward L D, Birney E, Crawford G E, Dekker J, Dunham I, Elnitski L L, Farnham P J, Feingold E A, Gerstein M, Giddings M C, Gilbert D M, Gingeras T R, Green E D, Guigo R, Hubbard T, Kent J, Lieb J D, Myers R M, Pazin M J, Ren B, Stamatoyannopoulos J A, Weng Z, White K P, Hardison R C (2014). Defining functional DNA elements in the human genome. Proc Natl Acad Sci USA, 111(17): 6131–6138
Kim D D, Kim T T, Walsh T, Kobayashi Y, Matise T C, Buyske S, Gabriel A (2004). Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res, 14(9): 1719–1725
Levanon E Y, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman Z Y, Shoshan A, Pollock S R, Sztybel D, Olshansky M, Rechavi G, Jantsch M F (2004). Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol, 22(8): 1001–1005
Lo P K, Zhang Y, Wolfson B, Gernapudi R, Yao Y, Duru N, Zhou Q (2016). Dysregulation of the BRCA1/long non-coding RNA NEAT1 signaling axis contributes to breast tumorigenesis. Oncotarget, (In press)
Lu Y, Li T, Wei G, Liu L, Chen Q, Xu L, Zhang K, Zeng D, Liao R (2016). The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol, 37(9): 11733–11741
Ma Y, Liu L, Yan F, Wei W, Deng J, Sun J (2016). Enhanced expression of long non-coding RNA NEAT1 is associated with the progression of gastric adenocarcinomas. World J Surg Oncol, 14 (1): 41
Mao Y S, Zhang B, Spector D L (2011). Biogenesis and function of nuclear bodies. Trends Genet, 27(8): 295–306
Mercer TR, Dinger ME, Mattick J S (2009). Long non-coding RNAs: insights into functions. Nature reviews, 10(3): 155–159
Naganuma T, Nakagawa S, Tanigawa A, Sasaki Y F, Goshima N, Hirose T (2012). Alternative 3'-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J, 31(20): 4020–4034
Nakagawa S, Naganuma T, Shioi G, Hirose T (2011). Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol, 193(1): 31–39
Nakagawa S, Shimada M, Yanaka K, Mito M, Arai T, Takahashi E, Fujita Y, Fujimori T, Standaert L, Marine J C, Hirose T (2014). The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development, 141(23): 4618–4627
Nasr R, Guillemin M C, Ferhi O, Soilihi H, Peres L, Berthier C, Rousselot P, Robledo-Sarmiento M, Lallemand-Breitenbach V, Gourmel B, Vitoux D, Pandolfi P P, Rochette-Egly C, Zhu J, de Thé H (2008). Eradication of acute promyelocytic leukemia-initiating cells through PML-RARA degradation. Nat Med, 14(12): 1333–1342
Peng Y, Yu S, Li H, Xiang H, Peng J, Jiang S (2014). MicroRNAs: emerging roles in adipogenesis and obesity. Cell Signal, 26(9): 1888–1896
Platani M, Lamond A I (2004). Nuclear organisation and subnuclear bodies. Prog Mol Subcell Biol, 35: 1–22
Poomsawat S, Sanguansin S, Punyasingh J, Vejchapipat P, Punyarit P (2016). Expression of cdk6 in head and neck squamous cell carcinoma. Clin Oral Investig, 20(1): 57–63
Prasanth K V, Prasanth S G, Xuan Z, Hearn S, Freier S M, Bennett C F, ZhangMQ, Spector D L (2005). Regulating gene expression through RNA nuclear retention. Cell, 123(2): 249–263
Prasanth K V, Spector D L (2007). Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev, 21(1): 11–42
Ricke WA, McPherson S J, Bianco J J, Cunha G R, Wang Y, Risbridger G P (2008). Prostatic hormonal carcinogenesis is mediated by in situ estrogen production and estrogen receptor alpha signaling. FASEB J, 22(5): 1512–1520
Sasaki Y T, Ideue T, Sano M, Mituyama T, Hirose T (2009). MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA, 106(8): 2525–2530
Schmitt A M, Chang H Y (2016). Long Noncoding RNAs in Cancer Pathways. Cancer Cell, 29(4): 452–463
Semenza G L (2010). Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene, 29(5): 625–634
Setlur S R, Mertz K D, Hoshida Y, Demichelis F, Lupien M, Perner S, Sboner A, Pawitan Y, Andrén O, Johnson L A, Tang J, Adami H O, Calza S, Chinnaiyan A M, Rhodes D, Tomlins S, Fall K, Mucci L A, Kantoff PW, StampferMJ, Andersson S O, Varenhorst E, Johansson J E, Brown M, Golub T R, Rubin M A (2008). Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst, 100(11): 815–825
Simon M D, Wang C I, Kharchenko P V, West J A, Chapman B A, Alekseyenko A A, Borowsky M L, Kuroda M I, Kingston R E (2011). The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA, 108(51): 20497–20502
Souquere S, Beauclair G, Harper F, Fox A, Pierron G (2010). Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell, 21(22): 4020–4027
Standaert L, Adriaens C, Radaelli E, Van Keymeulen A, Blanpain C, Hirose T, Nakagawa S, Marine JC (2014). The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA (New York, NY, 20(12): 1844–1849
Sunwoo H, Dinger M E, Wilusz J E, Amaral P P, Mattick J S, Spector D L (2009). MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res, 19(3): 347–359
Wang K C, Chang H Y (2011). Molecular mechanisms of long noncoding RNAs. Mol Cell, 43(6): 904–914
Wang P, Wu T, Zhou H, Jin Q, He G, Yu H, Xuan L, Wang X, Tian L, Sun Y, Liu M, Qu L (2016). Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J Exp Clin Cancer Res, 35 (1): 22
Waterston R H, Lindblad-Toh K, Birney E, Rogers J, Abril J F, Agarwal P, Agarwala R, Ainscough R, AlexanderssonM, An P, Antonarakis S E, Attwood J, Baertsch R, Bailey J, BarlowK, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, BrentMR, Brown D G, Brown S D, Bult C, Burton J, Butler J, Campbell R D, Carninci P, Cawley S, Chiaromonte F, Chinwalla A T, Church DM, Clamp M, Clee C, Collins F S, Cook L L, Copley R R, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty K D, Deri J, Dermitzakis E T, Dewey C, Dickens N J, Diekhans M, Dodge S, Dubchak I, Dunn D M, Eddy S R, Elnitski L, Emes R D, Eswara P, Eyras E, Felsenfeld A, Fewell G A, Flicek P, Foley K, FrankelWN, Fulton L A, Fulton R S, Furey T S, Gage D, Gibbs R A, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves T A, Green E D, Gregory S, Guigó R, Guyer M, Hardison R C, Haussler D, Hayashizaki Y, Hillier L W, Hinrichs A, Hlavina W, HolzerT, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe D B, Johnson L S, Jones M, Jones T A, Joy A, Kamal M, Karlsson E K, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent W J, Kirby A, Kolbe D L, Korf I, Kucherlapati R S, Kulbokas E J, Kulp D, Landers T, Leger J P, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott D R, Mardis E R, Matthews L, Mauceli E, Mayer J H, McCarthy M, McCombie W R, McLaren S, McLay K, McPherson J D, Meldrim J, Meredith B, Mesirov J P, Miller W, Miner T L, Mongin E, Montgomery K T, Morgan M, Mott R, Mullikin J C, Muzny D M, Nash W E, Nelson J O, Nhan M N, Nicol R, Ning Z, Nusbaum C, O’Connor M J, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin K H, Peterson J, Pevzner P, Plumb R, Pohl C S, Poliakov A, Ponce T C, Ponting C P, Potter S, Quail M, ReymondA, Roe B A, Roskin K M, Rubin E M, Rust A G, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz M S, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer J B, Slater G, Smit A, Smith D R, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson J P, Von Niederhausern A C, Wade C M, Wall M, Weber R J, Weiss R B, Wendl M C, West A P, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, WilliamsS, Wilson R K, Winter E, Worley K C, Wyman D, Yang S, Yang S P, Zdobnov E M, Zody M C, Lander E S, and the Mouse Genome Sequencing Consortium (2002). Initial sequencing and comparative analysis of the mouse genome. Nature, 420(6915): 520–562
West J A, Davis C P, Sunwoo H, Simon M D, Sadreyev R I, Wang P I, Tolstorukov M Y, Kingston R E (2014). The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell, 55(5): 791–802
Yamazaki T, Hirose T (2015). The building process of the functional paraspeckle with long non-coding RNAs. Front Biosci (Elite Ed), 7(1): 1–41
You J, Zhang Y, Liu B, Li Y, Fang N, Zu L, Li X, Zhou Q (2014). MicroRNA-449a inhibits cell growth in lung cancer and regulates long noncoding RNA nuclear enriched abundant transcript 1. Indian J Cancer, 51 (7 Suppl 3): e77–e81
Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L, Chen S, Li Y (2014). Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer, 14 (1): 693
Zhang Q, Chen C Y, Yedavalli V S, Jeang K T (2013). NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio, 4 (1): e00596–e12
Zhang Z, Carmichael G G (2001). The fate of dsRNA in the nucleus: a p54 (nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell, 106(4): 465–475
Zhen L, Yun-Hui L, Hong-Yu D, Jun M, Yi-Long Y (2016). Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol, 37(1): 673–683
Zhu J, Gianni M, Kopf E, Honore N, Chelbi-Alix M, Koken M, Quignon F, Rochette-Egly C, de The H (1999). Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor alpha (RARalpha) and oncogenic RARalpha fusion proteins. Proc Natl Acad Sci U S A, 96(26): 14807–14812
Zolotukhin A S, Michalowski D, Bear J, Smulevitch S V, Traish A M, Peng R, Patton J, Shatsky I N, Felber B K (2003). PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol Cell Biol, 23(18): 6618–6630
Acknowledgments
This work was supported by National Institutes of Health (NIH) R01 grants [grant numbers 5R01CA157779-03, 5R01CA163820-04] to Q.Z.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lo, PK., Wolfson, B. & Zhou, Q. Cellular, physiological and pathological aspects of the long non-coding RNA NEAT1. Front. Biol. 11, 413–426 (2016). https://doi.org/10.1007/s11515-016-1433-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11515-016-1433-z