Abstract
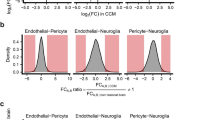
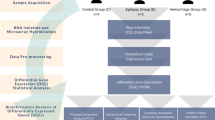
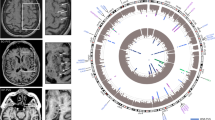
Brain senescence is associated with impaired endothelial barrier function, angiogenic and inflammatory activity, and propensity to brain hemorrhage. The same pathological changes occur in cerebral cavernous malformations (CCM), a genetic neurovascular anomaly. We hypothesized common transcriptomic and plasma cytokine signatures in the aging brain and CCM. We identified 320 genes [fold change ≥1.5; p < 0.05; false discovery rate (FDR) corrected] commonly dysregulated in the aging brain and CCM. Ontology and pathway analyses of the common differentially expressed genes were related to inflammation and extracellular matrix organization. Plasma levels of C-reactive protein and angiopoietin-2 were significantly greater in older compared to younger healthy non-CCM subjects and were also greater in CCM (Sporadic and Familial) subjects regardless of age (all: p < 0.05; FDR corrected). Plasma levels of vascular endothelial growth factor were significantly greater in older compared to younger subjects, in both healthy non-CCM and Sporadic-CCM groups (all: padj < 0.05). Plasma levels of vascular endothelial growth factor were also significantly greater in Familial-CCM cases with germ line mutations regardless of age (all: padj < 0.05) compared to both healthy non-CCM and Sporadic-CCM subjects. Brain white matter vascular permeability assessed by MRI followed the same pattern as vascular endothelial growth factor across all groups. In addition, quantitative susceptibility mapping of brain white matter, a measure of iron deposition, was increased in older compared to younger healthy non-CCM subjects. Genetic aberrations, plasma molecules, and imaging biomarkers in a well characterized Mendelian neurovascular disease may also be applicable in the aging brain.

Graphical abstract




Similar content being viewed by others
References
Akers A, al-Shahi Salman R, A. Awad I, Dahlem K, Flemming K, Hart B, et al. Synopsis of guidelines for the clinical management of cerebral cavernous malformations: consensus recommendations based on systematic literature review by the Angioma Alliance scientific advisory board clinical experts panel. Neurosurgery. 2017;80:665–80. https://doi.org/10.1093/neuros/nyx091.
Bottazzi B, Inforzato A, Messa M, Barbagallo M, Magrini E, Garlanda C, et al. The pentraxins PTX3 and SAP in innate immunity, regulation of inflammation and tissue remodelling. J Hepatol. 2016;64:1416–27. https://doi.org/10.1016/j.jhep.2016.02.029.
Chen L, Liu W, Wang P, Xue Y, Su Q, Zeng C, et al. Endophilin-1 regulates blood-brain barrier permeability via EGFR-JNK signaling pathway. Brain Res. 2015;1606:44–53. https://doi.org/10.1016/j.brainres.2015.02.032.
Colvin RA, Means TK, Diefenbach TJ, Moita LF, Friday RP, Sever S, et al. Synaptotagmin-mediated vesicle fusion regulates cell migration. Nat Immunol. 2010;11:495–502. https://doi.org/10.1038/ni.1878.
Diz AP, Carvajal-Rodriguez A, Skibinski DO. Multiple hypothesis testing in proteomics: a strategy for experimental work. Mol Cell Proteomics. 2011;10(M110):004374. https://doi.org/10.1074/mcp.M110.004374.
Elahy M, Jackaman C, Mamo J, Lam V, Dhaliwal SS, Giles C, et al. Blood-brain barrier dysfunction developed during normal aging is associated with inflammation and loss of tight junctions but not with leukocyte recruitment. Immun Ageing. 2015;12:2. https://doi.org/10.1186/s12979-015-0029-9.
Erdo F, Denes L, de Lange E. Age-associated physiological and pathological changes at the blood-brain barrier: a review. J Cereb Blood Flow Metab. 2017;37:4–24. https://doi.org/10.1177/0271678X16679420.
Girard R, Zeineddine HA, Fam MD, Mayampurath A, Cao Y, Shi C, et al. Plasma biomarkers of inflammation reflect seizures and hemorrhagic activity of cerebral cavernous malformations. Transl Stroke Res. 2018a;9:34–43. https://doi.org/10.1007/s12975-017-0561-3.
Girard R, Zeineddine HA, Koskimäki J, Fam MD, Cao Y, Shi C, et al. Plasma biomarkers of inflammation and angiogenesis predict cerebral cavernous malformation symptomatic hemorrhage or lesional growth. Circ Res. 2018b;122:1716–21. https://doi.org/10.1161/CIRCRESAHA.118.312680.
Golden MJ, Morrison LA, Kim H, Hart BL. Increased number of white matter lesions in patients with familial cerebral cavernous malformations. Am J Neuroradiol. 2015;36:899–903. https://doi.org/10.3174/ajnr.A4200.
Gurung P, Fan G, Lukens JR, Vogel P, Tonks NK, Kanneganti TD. Tyrosine kinase SYK licenses MyD88 adaptor protein to instigate IL-1alpha-mediated inflammatory disease. Immunity. 2017;46:635–48. https://doi.org/10.1016/j.immuni.2017.03.014.
Hamada K, Oike Y, Takakura N, Ito Y, Jussila L, Dumont DJ, et al. VEGF-C signaling pathways through VEGFR-2 and VEGFR-3 in vasculoangiogenesis and hematopoiesis. Blood. 2000;96:3793–800. https://doi.org/10.1182/blood.V96.12.3793.h8003793_3793_3800.
Jenny Zhou H, et al. Endothelial exocytosis of angiopoietin-2 resulting from CCM3 deficiency contributes to cerebral cavernous malformation. Nat Med. 2016;22:1033–42. https://doi.org/10.1038/nm.4169.
Joseph J, Cole G, Head E, Ingram D. Nutrition, brain aging, and neurodegeneration. J Neurosci. 2009;29:12795–801. https://doi.org/10.1523/JNEUROSCI.3520-09.2009.
Koskimaki J, et al. Comprehensive transcriptome analysis of cerebral cavernous malformation across multiple species and genotypes. JCI insight. 2019;4:e126167. https://doi.org/10.1172/jci.insight.126167.
Krouwer VJ, Hekking LH, Langelaar-Makkinje M, Regan-Klapisz E, Post JA. Endothelial cell senescence is associated with disrupted cell-cell junctions and increased monolayer permeability. Vasc Cell. 2012;4:12. https://doi.org/10.1186/2045-824X-4-12.
Lahteenvuo J, Rosenzweig A. Effects of aging on angiogenesis. Circ Res. 2012;110:1252–64. https://doi.org/10.1161/CIRCRESAHA.111.246116.
Lampugnani MG. Endothelial cell-to-cell junctions: adhesion and signaling in physiology and pathology. Cold Spring Harb Perspect Med. 2012;2:a006528. https://doi.org/10.1101/cshperspect.a006528.
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3. https://doi.org/10.1093/bioinformatics/bts034.
Lopez-Ramirez MA, Fonseca G, Zeineddine HA, Girard R, Moore T, Pham A, et al. Thrombospondin1 (TSP1) replacement prevents cerebral cavernous malformations. J Exp Med. 2017;214:3331–46. https://doi.org/10.1084/jem.20171178.
Lopez-Ramirez MA, Pham A, Girard R, Wyseure T, Hale P, Yamashita A, et al. Cerebral cavernous malformations form an anticoagulant vascular domain in humans and mice. Blood. 2019;133:193–204. https://doi.org/10.1182/blood-2018-06-856062.
Lu J, Marjon KD, Marnell LL, Wang R, Mold C, Du Clos TW, et al. Recognition and functional activation of the human IgA receptor (FcalphaRI) by C-reactive protein. PNAS. 2011;108:4974–9. https://doi.org/10.1073/pnas.1018369108.
Lyne SB, Girard R, Koskimäki J, Zeineddine HA, Zhang D, Cao Y, et al. Biomarkers of cavernous angioma with symptomatic hemorrhage. JCI insight. 2019;4:e128577. https://doi.org/10.1172/jci.insight.128577.
Mikati AG, Tan H, Shenkar R, Li L, Zhang L, Guo X, et al. Dynamic permeability and quantitative susceptibility: related imaging biomarkers in cerebral cavernous malformations. Stroke. 2014;45:598–601. https://doi.org/10.1161/STROKEAHA.113.003548.
Mikati AG, Khanna O, Zhang L, Girard R, Shenkar R, Guo X, et al. Vascular permeability in cerebral cavernous malformations. J Cereb Blood Flow Metab. 2015;35:1632–9. https://doi.org/10.1038/jcbfm.2015.98.
Mineo C, Gormley AK, Yuhanna IS, Osborne-Lawrence S, Gibson LL, Hahner L, et al. FcgammaRIIB mediates C-reactive protein inhibition of endothelial NO synthase. Circ Res. 2005;97:1124–31. https://doi.org/10.1161/01.RES.0000194323.77203.fe.
Poggesi A, Pasi M, Pescini F, Pantoni L, Inzitari D. Circulating biologic markers of endothelial dysfunction in cerebral small vessel disease: a review. J Cereb Blood Flow Metab. 2016;36:72–94. https://doi.org/10.1038/jcbfm.2015.116.
Rajani RM, et al. Reversal of endothelial dysfunction reduces white matter vulnerability in cerebral small vessel disease in rats. Sci Transl Med. 2018;10:eaam9507. https://doi.org/10.1126/scitranslmed.aam9507.
Rigamonti N, Kadioglu E, Keklikoglou I, Wyser Rmili C, Leow CC, De Palma M. Role of angiopoietin-2 in adaptive tumor resistance to VEGF signaling blockade. Cell Rep. 2014;8:696–706. https://doi.org/10.1016/j.celrep.2014.06.059.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. LIMMA powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. https://doi.org/10.1093/nar/gkv007.
Shi C, Shenkar R, Kinloch A, Henderson SG, Shaaya M, Chong AS, et al. Immune complex formation and in situ B-cell clonal expansion in human cerebral cavernous malformations. J Neuroimmunol. 2014;272:67–75. https://doi.org/10.1016/j.jneuroim.2014.04.016.
Stockton RA, Shenkar R, Awad IA, Ginsberg MH. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J Exp Med. 2010;207:881–96. https://doi.org/10.1084/jem.20091258.
Tan H, Liu T, Wu Y, Thacker J, Shenkar R, Mikati AG, et al. Evaluation of iron content in human cerebral cavernous malformation using quantitative susceptibility mapping. Investig Radiol. 2014;49:498–504. https://doi.org/10.1097/RLI.0000000000000043.
Tan H, Zhang L, Mikati AG, Girard R, Khanna O, Fam MD, et al. Quantitative susceptibility mapping in cerebral cavernous malformations: clinical correlations. Am J Neuroradiol. 2016;37:1209–15. https://doi.org/10.3174/ajnr.A4724.
Van Selst M, Jolicoeur P. A solution to the effect of sample size on outlier elimination. Q J Exp Psychol. 1994;47:631–50. https://doi.org/10.1080/14640749408401131.
Whitehead KJ, Chan AC, Navankasattusas S, Koh W, London NR, Ling J, et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat Med. 2009;15:177–84. https://doi.org/10.1038/nm.1911.
Wimo A, Guerchet M, Ali GC, Wu YT, Prina AM, Winblad B, et al. The worldwide costs of dementia 2015 and comparisons with 2010. Alzheimers Dement. 2017;13:1–7. https://doi.org/10.1016/j.jalz.2016.07.150.
Winkler MS, Rissiek A, Priefler M, Schwedhelm E, Robbe L, Bauer A, et al. Human leucocyte antigen (HLA-DR) gene expression is reduced in sepsis and correlates with impaired TNFalpha response: a diagnostic tool for immunosuppression? PLoS One. 2017;12:e0182427. https://doi.org/10.1371/journal.pone.0182427.
Yokoyama JS, Wang Y, Schork AJ, Thompson WK, Karch CM, Cruchaga C, et al. Association between genetic traits for immune-mediated diseases and Alzheimer disease. JAMA Neurol. 2016;73:691–7. https://doi.org/10.1001/jamaneurol.2016.0150.
Zeineddine HA, Girard R, Saadat L, Shen L, Lightle R, Moore T, et al. Phenotypic characterization of murine models of cerebral cavernous malformations. Lab Investig. 2019;99:319–30. https://doi.org/10.1038/s41374-018-0030-y.
Zhou Z, Tang AT, Wong WY, Bamezai S, Goddard LM, Shenkar R, et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature. 2016;532:122–6. https://doi.org/10.1038/nature17178.
Availability of data and material
The raw sequencing data for the human laser-captured lesional CCM NVUs used in this study are freely available in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database and are accessible through GEO series accession number GSE130176 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
Funding
The National Institutes of Health R21NS087328 and P01 NS092521 grants, the University of Chicago Comprehensive Cancer Center (P30 CA14599), the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1 TR000430), the William and Judith Davis Fund in Neurovascular Surgery Research at the University of Chicago, the Grossman Institute for Neuroscience, Quantitative Biology and Human Behavior at the University of Chicago, and the Be Brave for Life Foundation to IAA; the Safadi Program at the University of Chicago Translational Fellowship to RG and pilot grants to SPP and DZ; the Sigrid Juselius Foundation, Emil Aaltonen foundation and Maud Kuistila foundation to JK; and The American Association of Neurological Surgeons/Congress of Neurological Surgeons Joint Cerebrovascular Section Robert J. Dempsey MD Cerebrovascular Research Grant to SPP.
Author information
Authors and Affiliations
Contributions
JK was responsible for the figures, data acquisition (imaging, transcriptome, and plasma), data analysis (imaging, transcriptome, and plasma), data interpretation (imaging, transcriptome, and plasma), and writing; SP did data collection (transcriptome), data analysis (transcriptome), data interpretation (transcriptome), and writing; YL was responsible for the figures, data analysis (transcriptome), data interpretation (transcriptome); SR did data collection (imaging, transcriptome, and plasma), data interpretation (imaging, transcriptome, and plasma), and writing; AS did data collection (imaging, transcriptome, and plasma), data interpretation (imaging, transcriptome, and plasma), and writing; DZ did data collection (transcriptome and plasma); JCP did data collection (imaging and plasma); RL did data collection (transcriptome and plasma); TM did data collection (transcriptome and plasma); SL did data collection (transcriptome and plasma), data interpretation (imaging, transcriptome, and plasma), and writing; AS was responsible for the design and imaging data acquisition; KP was responsible for the design and data acquisition; YC did data analysis; RS did the study design; AD did imaging data analysis and interpretation; NH did imaging data acquisition and analysis; GAC was responsible for the design and imaging data acquisition; TC did the design and imaging data acquisition; RG did the study design, figures, imaging data acquisition, transcriptome data collection, plasma data collection, imaging data analysis, transcriptome data analysis, plasma data analysis, imaging data interpretation, transcriptome data interpretation, plasma data interpretation, and writing; and IA did the study design, imaging data interpretation, transcriptome data interpretation, plasma data interpretation, and writing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent to participate and for publication
Informed consent was obtained from all individual participants included in the study (AURA IRB: #13–0448).
Code availability
Not applicable
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Koskimäki, J., Polster, S.P., Li, Y. et al. Common transcriptome, plasma molecules, and imaging signatures in the aging brain and a Mendelian neurovascular disease, cerebral cavernous malformation. GeroScience 42, 1351–1363 (2020). https://doi.org/10.1007/s11357-020-00201-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-020-00201-4