Abstract
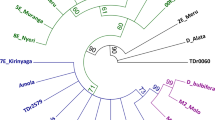
Wild cherry (Prunus avium L.) is a plant widely distributed around the world that possesses a great economic value. Indeed, it represents both a fruit source and one of the most important European hardwood species. For this reason, in order to obtain information about Turkish authoctonous P. avium germplasm and favor wood management policies and preservation strategies, we investigated the phylogenetic relationships existing among seven wild cherry populations (a total of 139 individuals) located in Northern Turkey using DNA barcoding. For each specimen, in detail, we sequenced a nuclear one (ITS) and two plastidial (trnH-psbA and matK) genes, to identify nucleotide polymorphisms, hypervariable genetic regions, and mutation events. Applying neighbor-joining method and genetic structure analysis, a high rate of crossbreeding among stands was revealed, except for one population (Gölcük) whose molecular profile was less similar to the others. In general, we observed that ITS was the most informative marker, suggesting it as a good candidate for P. avium intraspecific study. We conclude that DNA barcode technique, usually applied for species identification, may be also used as a scientific tool for the detection of plant biodiversity at population level.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Alvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol 29(3):417–434
Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann Missouri Bot Gard 82:247–277
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2004) GenBank: update. Nucleic Acids Res 32(Database issue):D23–D36
Bortiri E, Oh SH, Jiang J, Baggett S, Granger A, Weeks C, Parfitt DE (2001) Phylogeny and systematics of Prunus (Rosaceae) as determined by sequence analysis of ITS and the chloroplast trnL-trnF spacer DNA. Syst Bot 26(4):797–807
Bruni I, De Mattia F, Galimberti A, Galasso G, Banfi E, Casiraghi M, Labra M (2010) Identification of poisonous plants by DNA barcoding approach. Int J Legal Med 124(6):595–603
Bruni I, De Mattia F, Martellos S, Galimberti A, Savadori P, Casiraghi M, Labra M (2012) DNA barcoding as an effective tool in improving a digital plant identification system: a case study for the area of Mt Valerio, Trieste (NE Italy). PLoS One 7(9):e43256
Casiraghi M, Labra M, Ferri E, Galimberti A, De Mattia F (2010) DNA barcoding: a six-question tour to improve users’ awareness about the method. Brief Bioinform 11(4):440–453
Chiej R (1984) MacDonald encyclopedia of medicinal plants, London
De Rogatis A, Ferrazzini D, Ducci F, Guerri S, Carnevale S, Belletti P (2013) Genetic variation in Italian wild cherry (Prunus avium L) as characterized by nSSR markers. Forestry 86(3):391–400
Dong W, Liu J, Yu J, Wang L, Zhou S (2012) Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One 7(4):e35071
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno. Conserv Genet Res 4:359–361
Ercisli S (2004) A short review of the fruit germplasm resources of Turkey. Genet Resour Crop Evol 51(4):419–435
Erickson DL, Spouge J, Resch A, Weigt LA, Kress JW (2008) DNA barcoding in land plants: developing standards to quantify and maximize success. Taxon 57(4):1304–1316
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fazekas AJ (2008) Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS One 3:2802
Felsenstein J (1989) PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166
Felsenstein J (2005) PHYLIP (phylogeny inference package) version 3.6
Fernandez i, Marti A, Athanson B, Koepke T, Font i, Forcada C, Dhingra A, Oraguzie N (2012) Genetic diversity and relatedness of sweet cherry (Prunus avium L.) cultivars based on single nucleotide polymorphic markers. Front Plant Sci 3:116
Ganopoulos IV, Kazantzis K, Chatzicharisis I, Karayiannis I, Tsaftaris AS (2011) Genetic diversity, structure and fruit trait associations in Greek sweet cherry cultivars using microsatellite based (SSR/ISSR) and morpho-physiological markers. Euphytica 181(2):237–251
Gere J, Yessoufou K, Daru BH, Mankga LT, Maurin O, van der Bank M (2013) Incorporating trnH-psbA to the core DNA barcodes improves significantly species discrimination within southern African Combretaceae. ZooKeys 365:129–147
Gismondi A, Rolfo MF, Leonardi D, Rickards O, Canini A (2012) Identification of ancient Olea europaea L and Cornus mas L by DNA barcoding. C R Biol 335(7):472–479
Gismondi A, Fanali F, Labarga JMM, Caiola MG, Canini A (2013) Crocus sativus L genomics and different DNA barcode applications. Plant Syst Evol 299(10):1859–1863
Gismondi A, Di Marco G, Delorenzo M, Canini A (2015) Upgrade of Castanea sativa (mill) genetic resources by sequencing of barcode markers. J Genet 94(3):519–524
Gismondi A, Di Marco G, Martini F, Sarti L, Crespan M, Martínez-Labarga C, Rickards O, Canini A (2016) Grapevine carpological remains revealed the existence of a Neolithic domesticated Vitis vinifera L specimen containing ancient DNA partially preserved in modern ecotypes. J Archaeol Sci 69:75–84
Grieve A (1984) Modern herbal. Penguin
Group CBOL Plant Working et al (2009) A DNA barcode for land plants. Proc Natl Acad Sci U S A 106(31):12794–12797
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT Nucl Acids Symp Ser 41:95–98
Hilu KW, Alice LA, Liang H (1999) Phylogeny of Poaceae inferred from matK sequences. Ann Mo Bot Gard 86:835–851
Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS One 6(5):1–13
Hoveka LN, van der Bank M, Boatwright JS, Bezeng BS, Yessoufou K (2016) The noncoding trnH-psbA spacer, as an effective DNA barcode for aquatic freshwater plants, reveals prohibited invasive species in aquarium trade in South Africa. S Afr J Mar Sci 102:208–216
Jarni K, De Cuyper B, Brus R (2012) Genetic variability of wild cherry (Prunus avium L) seed stands in Slovenia as revealed by nuclear microsatellite loci. PLoS One 7(7):1–5
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kocyan A, de Vogel EF, Conti E, Gravendeel B (2008) Molecular phylogeny of Aerides (Orchidaceae) based on one nuclear and two plastid markers: a step forward in understanding the evolution of the Aeridinae. Mol Phylogenet Evol 48(2):422–443
Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One 2(6):e508
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci U S A 102(23):8369–8374
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Lahaye R (2008) DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci U S A 105:2923–2928
Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S (2015) Plant DNA barcoding: from gene to genome. Biol Rev 90(1):157–166
Mohanty A, Martin JP, Aguinagalde I (2001) Chloroplast DNA study in wild populations and some cultivars of Prunus avium L. Theor Appl Genet 103(1):112–117
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comp Appl Biol Sci 12:357–358
Pang X, Song J, Zhu Y, Xu H, Huang L, Chen S (2011) Applying plant DNA barcodes for Rosaceae species identification. Cladistics 27(2):165–170
Perazzini R, Leonardi D, Ruggeri S, Alesiani D, D’Arcangelo G, Canini A (2008) Characterization of Phaseolus vulgaris L landraces cultivated in Central Italy. Plant Foods Hum Nutr 63(4):211–218
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Russell K (2003) Technical Guidelines for genetic conservation and use for wild cherry (Prunus avium), EUFORGEN, International Plant Genetic Resources Institute Rome Italy 1–6. http://www.euforgen.org/fileadmin/templates/euforgen.org/upload/Publications/Technical_guidelines/859_Technical_guidelines_for_genetic_conservation_and_use_for_Wild_cherry__Prunus_avium_.pdf. Accessed 10 Aug 2018
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Santos C, Pereira F (2018) Identification of plant species using variable length chloroplast DNA sequences. Forensic Sci Int Genet 36:1–12
Savolainen V, Cowan RS, Vogler AP, Roderick GK, Lane R (2005) Towards writing the encyclopaedia of life: an introduction to DNA barcoding. Philos Trans R Soc Lond Ser B Biol Sci 360(1462):1805–1811
Scaltsoyiannes A, Tsoulpha P, Iliev I, Theriou K, Tsaktsira M, Mitras D, Karanikas C, Mahmout S, Christopoulos V, Scaltsoyiannes V, Zaragotas D, Tzouvara A (2009) Vegetative propagation of ornamental genotypes of Prunus avium L. Prop Ornam Plant 9:198–206
Scholz H, Scholz I (1995) Prunoideae. In: Hegi G (ed) Illustrierte Flora von Mitteleuropa, 2nd edn. Blackwell Wissenschafts-Verlag, Berlin, pp 446–510
Shi S, Li J, Sun J, Yu J, Zhou S (2013) Phylogeny and classification of Prunus sensu lato (Rosaceae). J Integr Plant Biol 55(11):1069–1079
Tavaud M, Zanetto A, Santi F, Dirlewanger E (2001) Structuration of genetic diversity in cultivated and wild cherry trees using AFLP markers. Acta Hortic 263–269
Theodoridis S, Stefanaki A, Tezcan M, Aki C, Kokkini S, Vlachonasios KE (2012) DNAbarcoding in native plants of the Labiatae (Lamiaceae) family from Chios Island (Greece) and the adjacent Çesme-Karaburun Peninsula (Turkey). Mol Ecol Resour 12:620–633
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vaughan SP, Cottrell JE, Moodley DJ, Connolly T, Russell K (2007) Clonal structure and recruitment in British wild cherry (Prunus avium L). For Ecol Manag 242(2–3):419–430
Wang W, Wu Y, Yan Y, Ermakova M, Kerstetter R, Messing J (2010) DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol 10(1):205
Welk E, de Rigo D, Caudullo G (2016) Prunus avium in Europe: distribution, habitat, usage and threats. In: San-Miguel-Ayanz J, de Rigo D, Caudullo G, Houston Durrant T, Mauri A (eds) European Atlas of Forest Tree Species, 1st edn, Luxembourg, p e01491d
Yaman B (2003) Yabani kiraz (Cerasus avium (L.) Moench). GÜ-Orman Fakültesi Dergisi 3(1):114–122
Yu J, Xue JH, Zhou SL (2011) New universal matK primers for DNA barcoding angiosperms. J Syst Evol 49(3):176–181
Acknowledgments
The authors want to thank Prof. Ahu Altınkut Uncuoğlu and Dr. Ezgi Çabuk Şahin for their great contribution in data analysis and Miss Sophie Gart who revised the English form of this manuscript.
Data archiving statement
Genetic data were registered in GenBank database. ID number of each deposited sequence, with relative details, was reported in Supplemental Material - Table S1 and copied below.
Population | trnH-psbA | matK | ITS |
Abant Population (A) | MF431607 | MF431608 | MF431606 |
Molla Fenari Population (B) | MF590194 | MF590188 | MF590200 |
Yedigöller Population (C) | MF590195 | MF590189 | MF590201 |
Düzce Population (D) | MF590197 | MF590190 | MF590202 |
Melen Population (E) | MF590196 | MF590191 | MF590203 |
Kefken Population (F) | MF590198 | MF590192 | MF590204 |
Gölcük Population (G) | MF590199 | MF590193 | MF590205 |
Author information
Authors and Affiliations
Contributions
AG, AC YOC, and EV designed research; SGU performed research; BUK carried out the sampling; EV authorized the sampling; SGU, AG, and GDM analyzed data; SGU and AG wrote the paper; AC provided financial support; all authors edited, revised, and provided comments to the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by E. Dirlewanger
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key message
Application of DNA barcoding at intraspecific level, to investigate phylogenetic relationships existing among Turkish wild cherry populations, increase genetic knowledge of this species, and promote its conservation and breeding programs
Angelo Gismondi and Antonella Canini are co-last authors of this manuscript.
Electronic supplementary material
ESM 1
(DOCX 4367 kb)
Rights and permissions
About this article
Cite this article
Ünsal, S.G., Çiftçi, Y.Ö., Eken, B.U. et al. Intraspecific discrimination study of wild cherry populations from North-Western Turkey by DNA barcoding approach. Tree Genetics & Genomes 15, 16 (2019). https://doi.org/10.1007/s11295-019-1323-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-019-1323-z