Abstract
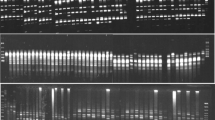
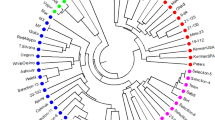
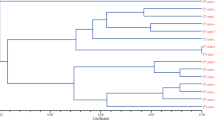
Knowledge of pistachio genetic diversity is necessary for the formulation of appropriate management strategies for the conservation of these species. We analysed amplified fragment length polymorphisms in a total of 216 pistachio accessions, which included seven populations from three wild species (Pistacia vera, Pistacia khinjuk and Pistacia atlantica subsp. kurdica) and most of the important cultivars from Iran, together with some foreign cultivars. High levels of genetic diversity were detected within the Iranian cultivars, and they showed a clear separation from foreign cultivars, as revealed by unweighted pair group method with arithmetic averaging and supported by analysis of molecular variance. The lowest amount of polymorphism was observed in P. atlantica subsp. kurdica, which showed the lowest number of total bands as compared to the other species. This revealed strong genetic erosion of P. atlantica subsp. kurdica, which reflected a severe decline in habitat and over-exploitation. Based on these findings, strategies are proposed for the genetic conservation and management of pistachio species and cultivars.







Similar content being viewed by others
References
Angiolillo A, Mencuccini M, Baldoni L (1999) Olive genetic diversity assessed using amplified fragment length polymorphisms. Theor Appl Genet 98:411–421
Bahsa AI, Padulosi S, Chabane K (2007) Genetic diversity of Syrian pistachio (Pistacia vera L.) varieties evaluated by AFLP markers. Gene Resour Crop Evol 54:1807–1816, doi:10.1007/s10722-006-9202-5
Esmail-pour A (2001) Distribution, use and conservation of pistachio in Iran. In: Padulosi S, Hadj-Hassan A (eds) In towards a comprehensive documentation and use of pistacia genetic diversity in central and West Asia, North Africa and Europe. Report of the IPGRI workshop, 14–17 December 1998, Ibrid, Jordan. IPGRI, Rome, Italy
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
FAO (2006) FAOSTAT database. http://apps.fao.org/page/form?collection= Production.Crops.Primary&Domain=Production&servlet=1&language=EN&hostname=apps.fao.org&version=default
Golan-Goldhirsh A, Barazani O, Wang ZS, Khadkal DK, Saunders JA, Kostiukovsky V, Rowland LJ (2004) Genetic relationships among Mediterranean Pistacia species evaluated by RAPD and AFLP markers. Plant Systemat Evol 246:9–18
Gower JC (1966) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338
Joret C (1976) Les Plantes dansL'antiquité et au moyen âge; histoire, usages et symbolisme. Slatkine Reprints, Genève. Reprinted from the book first published in 1897–1904
Hartl L, Seefleder S (1998) Diversity of selected Hop cultivars detected by fluorescent AFLPs. Theo Appl Gene 96:112–116
Hormaza JI, Wünsch A (2007) Pistachio. In: Kole C (ed) Genome mapping and molecular breeding in plants, fruits and nuts, vol 4. Springer, New York, USA, pp 243–251
Hormaza JI, Dollo L, Polito VS (1994) Determination of relatedness and geographic movements of Pistacia vera (Pistachio; Anacardiaceae) germplasm by RAPD analysis. Econ Bot 48:349–358
Kafkas S (2006) Phylogenetic analysis of the genus Pistacia by AFLP markers. Plant Syst Evol 262:113–124
Kafkas S, Perl-Treves R (2001) Morphological and molecular phylogeny of Pistacia species in Turkey. Theo Appl Gene 102:908–915
Kafkas S, Perl-Treves R (2002) Interspecific relationships in Pistacia based on RAPD fingerprinting. Hort Sci 37:168–171
Kafkas S, Kaska A, Wassimi AN, Padulosi S (2006a) Molecular characterisation of Afghan pistachio accessions by amplified fragment length polymorphisms (AFLPs). J Hort Sci Biotechnol 81:864–868
Kafkas S, Ozkan H, Erol Ak B, Acar I, Alti HS (2006b) Detecting DNA polymorphism and genetic diversity in a wide pistachio germplasm: comparison of AFLP, ISSR, and RAPD marker. J Am Soc Hort Sci 131:522–529
Katsiotis A, Hagidimitriou M, Drossoul A, Pontikis C, Loukas M (2003) Genetic relationships among species and cultivars of Pistacia using PAPDs and AFLPs. Euphytica 132:279–286
Kayimov AK, Sultanov RA, Chernova GM (2001) Pistacia in Central Asia. In: Padulosi S, Hadj-Hassan A (eds) Project on underutilizes Mediterranean species. Pistacia: towards a comprehensive documentation of distribution and use of its genetic diversity in Central & West Asia, North Africa and Mediterranean Europe. IPGRI, Rome, Italy
Khatamsaz M (1988) Flora of Iran No. 30: Anacardiaceae. Research Institute of Forests and Rangelands, Tehran, Iran
Maggs DH (1973) Genetic resources in pistachio. Plant Gene Resour Newsle 29:7–15
Manubens A, Lobos S, Jadue Y, Toro M, Messina R, Lladser M, Seelenfrund D (1999) DNA isolation and AFLP fingerprinting of nectarine and peach varieties (Prunus persica). Mol Biol Rep 17:255–267
Maughan PJ, Saghai Maroof MA Buss GR, Huestis GM (1996) Amplified fragment length polymorphism (AFLP) in soybean: species diversity, inheritance, and neaisogenic line analysis. Theo Appl Gene 93:392–401
Milligan BG, Leebens-Mack J, Strand AE (1994) Conservation genetics: beyond the maintenance of marker diversity. Mol Ecol 12:844–855
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Li W (1979) Mathematical model for study genetic variation in terms of restriction endonucleases. Proc Nati Acad Sci USA 74:5267–5273
Parfitt DE (1995) Pistachio cultivars. In: Ferguson L (ed) Pistachio production. University of California, Davis, pp 43–46
Peakal R, Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (Microsatellites) markers for germplasm analysis. Mol Breed 2:225–235
Rohlf JF (2004) NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.11. Exeter, Setauket, NY
Russell JR, Weber JC, Booth A, Powell W, Sotelo-Montes C, Dawson IK (1999) Genetic variation of Calycophyllum spruceanum in the Peruvian Amazon basin, revealed by amplified fragment length polymorphism (AFLP) analysis. Mol Ecol 8:199–204
Schneider S, Roessli D, Excoffier L (2000) Arlequin: A software for population genetics data analysis version 2.000 genetics and biometry laboratory, dept. of anthropology. University of Geneva, Switzerland
Sensi E, Vignani R, Rhode W, Biricolti S (1996) Characterization of genetic biodiversity with Vitis vinifera L. sangiovese and colorino genotypes by AFLP and ISTR DNA marker technology. Vitis 35:183–188
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tous J, Fergusen L (1996) Mediterranean fruits. In: Janick J (ed) Progress in new crops. ASHS, Arlington, VA
Vos P, Hogers R, Bleeker M, Reijans M, van der Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wang YH, Thomas CE, Dean RA (1997) A genetic map of melon (Cucumis melo L.) based on amplified fragment length polymorphism (AFLP) Markers. Theo Appl Gene 95:791–798
Whitehouse WE, Stone CL (1941) Some aspects of dichogamy and pollination in pistachio. Am Soc Hort Sci 39:95–100
Winfield MO, Arnold GM, Cooper F, Le Ray M, White J, Karp A, Edwards KJ (1998) A study of genetic diversity in Populus nigra subsp. betulifolia in The upper seven area of The UK using AFLP markers. Mol Ecol 7:3–10
Xu RQ, Tomooka N, Vaughan DA (2000) AFLP markers for characterizing the Azuki Bean complex. Crop Sci 40:808–815
Zohary M (1952) A monographical study of the genus Pistacia. Palest J Bot Jerus Ser 5:187–228
Zohary D (1996) The genus Pistacia L. In: Padulosi S, Caruso T, Barone E (eds) Taxonomy, distribution, conservation and uses of Pistacia genetic resources. IPGRI, Palermo, Italy, pp 1–11
Acknowledgments
This study was supported by the Agricultural Biotechnology Research Institute of Iran. The authors would like to thank Dr. Hasan Maddah Arefy from the Research Institute of Forests and Rangelands of Iran for the photographs of pistachio. They would also like to thank Dr A. A. Javanshah (Director General of the Iranian Pistachio Research Institute), in particular, for his contribution to the collection of plant material.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shanjani, P.S., Mardi, M., Pazouki, L. et al. Analysis of the molecular variation between and within cultivated and wild Pistacia species using AFLPs. Tree Genetics & Genomes 5, 447–458 (2009). https://doi.org/10.1007/s11295-008-0198-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-008-0198-1