Abstract
In the last two decades, there has been increasing evidence supporting the role of the intestinal microbiota in health and disease, as well as the use of probiotics to modulate its activity and composition. Probiotic bacteria selected for commercial use in foods, mostly lactic acid bacteria and bifidobacteria, must survive in sufficient numbers during the manufacturing process, storage, and passage through the gastro-intestinal tract. They have several modes of action and it is crucial to unravel the mechanisms underlying their postulated beneficial effects. To track their survival and persistence, and to analyse their interaction with the gastro-intestinal epithelia it is essential to discriminate probiotic strains from endogenous microbiota. Fluorescent reporter proteins are relevant tools that can be exploited as a non-invasive marker system for in vivo real-time imaging in complex ecosystems as well as in vitro fluorescence labelling. Oxygen is required for many of these reporter proteins to fluoresce, which is a major drawback in anoxic environments. However, some new fluorescent proteins are able to overcome the potential problems caused by oxygen limitations. The current available approaches and the benefits/disadvantages of using reporter vectors containing fluorescent proteins for labelling of bacterial probiotic species commonly used in food are addressed.


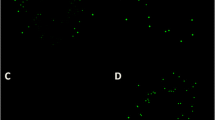
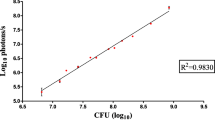
Image from Landete et al. (2015)
Similar content being viewed by others
References
Allison GE, Klaenhammer TR (1996) Functional analysis of the gene encoding immunity to lactacin F, lafI, and its use as a Lactobacillus-specific, food-grade genetic marker. Appl Environ Microbiol 62:4450–4460
Arqués JL, Hautefort I, Ivory K, Bertelli E, Regoli M, Clare S, Hinton JC, Nicoletti C (2009) Salmonella induces flagellin- and MyD88-dependent migration of bacteria-capturing dendritic cells into the gut lumen. Gastroenterology 137:579–587
Baird GS, Zacharias DA, Tsien RY (2000) Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA 97:11984–11989
Berlec A, Zavrsnik J, Butinar M, Turk B, Strukelj B (2015) In vivo imaging of Lactococcus lactis, Lactobacillus plantarum and Escherichia coli expresing infrared protein in mice. Microb Cell Fact 14:181
Bevis BJ, Glick BS (2002) Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat Biotechnol 20:83–87
Bongaerts RJM, Hautefort I, Sidebotham JM, Hinton JCD (2002) Green fluorescent protein as a marker for conditional gene expression in bacterial cells. Methods Enzymol 358:43–66
Brancaccio VF, Zhurina DS, Riedel CU (2013) Tough nuts to crack: site-directed mutagenesis of bifidobacteria remains a challenge. Bioengineered 4:197–202
Campelo AB, Roces C, Mohedano LM, López P, Rodríguez A, Martínez B (2014) A bacteriocin gene cluster able to enhance plasmid maintenance in Lactococcus lactis. Microb Cell Fact 13:77
Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC (1994) Green fluorescent protein as a marker for gene expression. Science 263:802–805
Chapman S, Faulkner C, Kaiserli E, Garcia-Mata C, Savenkov EI, Roberts AG, Oparka KJ, Christie JM (2008) The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc Natl Acad Sci USA 105:20038–20043
Chudakov DM, Lukyanov S, Lukyanov KA (2005) Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol 23:605–613
Coombes JL, Robey EA (2010) Dynamic imaging of host-pathogen interactions in vivo. Nat Rev Immunol 10:353–364
Cronin M, Sleator RD, Hill C, Fitzgerald GF, van Sinderen D (2008) Development of a luciferase-based reporter system to monitor Bifidobacterium breve UCC2003 persistence in mice. BMC Microbiol. doi:10.1186/1471-2180-8-161
Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY (1995) Understanding, improving and using green fluorescent proteins. Trends Biochem Sci 20:448–455
Cubitt AH, Wollenweber LA, Hein R (1999) Understanding structure–function relationships in the Aequorea victoria green fluorescent protein. Methods Cell Biol 58:19–30
Daniel C, Poiret S, Dennin V, Boutillier D, Pot B (2013) Bioluminescence imaging study of spatial and temporal persistence of Lactobacillus plantarum and Lactococcus lactis in living mice. Appl Environ Microbiol 79:1086–1094
Davidson MW, Campbell RE (2009) Engineered fluorescent proteins: innovations and applications. Nat Methods 6:713–717
Drepper T, Eggert T, Circolone F, Heck A, Krauss U, Guterl JK, Wendorff M, Losi A, Gartner W, Jaeger KE (2007) Reporter proteins for in vivo fluorescence without oxygen. Nat Biotechnol 25:443–445
Dupont L, Boizet-Bonhoure B, Coddeville M, Auvray F, Ritzenthaler P (1995) Characterization of genetic elements required for site-specific integration of Lactobacillus delbrueckii subsp. bulgaricus bacteriophage mv4 and construction of an integration-proficient vector for Lactobacillus plantarum. J Bacteriol 177:586–595
Ernst JF, Tielker D (2009) Responses to hypoxia in fungal pathogens. Cell Microbiol 11:183–190
Farnworth ER (2008) The evidence to support health claims for probiotics. J Nutr 138:1250S–1254S
Fernández de Palencia P, Nieto C, Acebo P, Espinosa M, Lopez P (2000) Expression of green fluorescent protein in Lactococcus lactis. FEMS Microbiol Lett 183:229–234
Fernández de Palencia PF, de la Plaza M, Mohedano ML, Martínez-Cuesta MC, Requena T, López P, Peláez C (2004) Enhancement of 2-methylbutanal formation in cheese by using a fluorescently tagged Lacticin 3147 producing Lactococcus lactis strain. Int J Food Microbiol 93:335–347
Fernández L, Marin M, Langa S, Martín R, Reviriego C, Fernández A, Olivares M, Xaus J, Rodríguez J (2004) A novel genetic label for detection of specific probiotic lactic acid bacteria. Food Sci Technol Int 10:101–108
Franz CMAP, Stiles ME, Schleifer KH, Holzapfel WH (2003) Enterococci in foods—a conundrum for food safety. Int J Food Microbiol 88:105–122
García-Cayuela T, de Cadiñanos LP, Mohedano ML, de Palencia PF, Boden D, Wells J, Peláez C, López P, Requena T (2012) Fluorescent protein vectors for promoter analysis in lactic acid bacteria and Escherichia coli. Appl Microbiol Biotechnol 96:171–181
Geoffroy MC, Guyard C, Quatannens B, Pavan S, Lange M, Mercenier A (2000) Use of green fluorescent protein to tag lactic acid bacterium strains under development as live vaccine vectors. Appl Environ Microbiol 66:383–391
Giepmans BNG (2008) Bridging Xuorescence microscopy and electron microscopy. Histochem Cell Biol 130:211–217
Giepmans BNG, Adams SR, Ellisman MH, Tsien RY (2006) The fluorescent toolbox for assessing protein location and function. Science 312:217–224
Giraffa G (2003) Functionality of enterococci in dairy products. Int J Food Microbiol 88:215–222
Gory L, Montel MC, Zagorec M (2001) Use of green fluorescent protein to monitor Lactobacillus sakei in fermented meat products. FEMS Microbiol Lett 194:127–133
Greer LF, Szalay AA (2002) Imaging of light emission from the expression of luciferases in living cells and organisms: a review. Luminescence 17:43–74
Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY (2001) Reducing the environmental sensitivity of yellow fluorescent protein: mechanism and applications. J Biol Chem 276:29188–29194
Grimm V, Gleinser M, Neu C, Zhurina D, Riedel CU (2014) Expression of fluorescent proteins in bifidobacteria for analysis of host-microbe interactions. Appl Environ Microbiol 80:2842–2850
Guinane CM, Piper C, Draper LA, O’Connor PM, Hill C, Ross RP, Cotter PD (2015) Impact of environmental factors on bacteriocin promoter activity in gut-derived Lactobacillus salivarius. Appl Environ Microbiol 81:7851–7859
Gurskaya NG, Fradkov AF, Pounkova NI, Staroverov DB, Bulina ME, Yanushevich YG, Labas YA, Lukyanov S, Lukyanov KA (2003) A colorless green fluorescent protein homologue from the non-fluorescent hydromedusa Aequorea coerulescens and its fluorescent mutants. Biochem J 373:403–408
Han X, Wang L, Li W, Li B, Yang Y, Yan H, Qu L, Chen Y (2015) Use of green fluorescent protein to monitor Lactobacillus plantarum in the gastrointestinal tract of goats. Braz J Microbiol 46:849–854
Hansen MC, Palmer RJ, Udsen C, White DC, Molin S (2001) Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383–1391
Hashemzadeh F, Rahimi S, Karimi Torshizi MA, Masoudi AA (2015) Usage of green fluorescent protein for tracing probiotic bacteria in alimentary tract and efficacy evaluation of different probiotic administration methods in broilers. J Agric Sci Technol 17:345–356
Heim R, Tsien RY (1996) Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol 6:178–182
Heim R, Prasher DC, Tsien RY (1994) Wavelength mutations and post-translational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA 91:12501–12504
Heim R, Cubitt AB, Tsien RY (1995) Improved green fluorescence. Nature 373:663–664
Heimbach JT (2008) Health-benefit claims for probiotic products. Clin Infect Dis 46:S122–S124
Hertel C, Ludwig W, Schleifer KH (1992) Introduction of silent mutations in a proteinase gene of Lactococcus lactis as a useful marker for monitoring studies. Syst Appl Microbiol 15:447–452
Hutchens M, Luker GD (2007) Applications of bioluminescence imaging to the study of infectious diseases. Cell Microbiol 9:2315–2322
Karasawa S, Araki T, Yamamoto-Hino M, Miyawaki A (2003) A green-emitting fluorescent protein from Galaxeidae coral and its monomeric version for use in fluorescent labeling. J Biol Chem 278:34167–34171
Karasawa S, Araki T, Nagi T, Mizuno H, Miyawaki A (2004) Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem J 381:307–312
Karimi S, Ahl D, Vagesjö E, Holm L, Phillipson M, Jonsson H, Ross S (2016) In vivo and in vitro detection of luminiscent and fluorescent Lactobacillus reuteri and application of red fluorescent protein mCherry for assesing plasmid persistence. PLoS ONE 11:e0151969
Kaushik JK, Kumar A, Duary RK, Mohanty AK, Grover S, Batish VK (2009) Functional and probiotic attributes of ana indigenous isolate of Lactobacillus plantarum. PLoS ONE 4:e8099
Klijn N, Weerkamp AH, de Vos WM (1995) Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl Environ Microbiol 61:2771–2774
Kremers GJ, Gilbert SG, Cranfill PJ, Davidson MW, Piston DW (2011) Fluorescent proteins at a glance. J Cell Sci 124:157–160
Kubota H, Senda S, Nomura N, Tokuda H, Uchiyama H (2008) Biofilm formation by lactic acid bacteria and resistance to environmental stress. J Biosci Bioeng 106:381–386
Lagendijk EL, Validov S, Lamers GEM, de Weert S, Bloemberg GV (2010) Genetic tools for tagging gram-negative bacteria with mCherry for visualization in vitro and in natural habitats, biofilm and pathogenicity studies. FEMS Microbiol Lett 305:81–90
Landete JM, Peirotén A, Rodríguez E, Margolles A, Medina M, Arqués JL (2014) Anaerobic green fluorescent protein as a marker of Bifidobacterium strains. Int J Food Microbiol 175:6–13
Landete JM, Langa S, Revilla C, Margolles A, Medina M, Arqués JL (2015) Use of anaerobic green fluorescent protein versus green fluorescent protein as reporter in lactic acid bacteria. Appl Microbiol Biotechnol 99:6865–6877
Larrainzar E, O’Gara F, Morrissey JP (2005) Applications of autofluorescent proteins for in situ studies in microbial ecology. Annu Rev Microbiol 59:257–277
Lee JH, O’Sullivan DJ (2010) Genomic insights into bifidobacteria. Microbiol Mol Biol Rev 74:378–416
Lillehaug D, Nes IF, Birkeland NK (1997) A highly efficient and stable systems for site-specific integration of genes and plasmids into the phage φLC3 attachment site (attB) of the Lactococcus lactis chromosome. Gene 188:129–136
Maguin E, Prévost H, Ehrlich D, Gruss A (1996) Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol 178:931–935
Manta C, Heupel E, Radulovic K, Rossini V, Garbi N, Riedel CU, Niess JH (2013) CX(3)CR1(+) macrophages support IL-22 production by innate lymphoid cells during infection with Citrobacter rodentium. Mucosal Immunol 6:177–188
Maruo T, Sakamoto M, Toda T, Benno Y (2006) Monitoring the cell number of Lactococcus lactis subsp cremoris FC in human feces by real-time PCR with strain-specific primers designed using the RAPD technique. Int J Food Microbiol 110:69–76
Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17:969–973
McAuliffe O, Hill C, Ross RP (2000) Identification and overexpression of ltnI, a novel gene which confers immunity to the two-component lantibiotic lacticin 3147. Microbiology 146:129–138
Miyawaki A (2005) Innovations in the imaging of brain functions using fluorescent proteins. Neuron 48:189–199
Miyawaki A, Griesbeck O, Heim R, Tsien RY (1999) Dyanmic and quantitative calcium measurements using improved cameleons. Proc Natl Acad Sci USA 96:2135–2140
Mohedano ML, Garcia-Cayuela T, Perez-Ramos A, Gaiser RA, Requena T, López P (2015) Construction and validation of a mCherry protein vector for promoter analysis in Lactobacillus acidophilus. J Ind Microbiol Biotechnol 42:247–253
Montenegro-Rodríguez C, Peirotén A, Sanchez-Jimenez A, Arqués JL, Landete JM (2015) Analysis of gene expression of bifidobacteria using as the reporter an anaerobic fluorescent protein. Biotechnol Lett 37:1405–1413
Mukherjee A, Schroeder CM (2015) Flavin-based fluorescent proteins: emerging paradigms in biological imaging. Curr Opin Biotechnol 31:16–23
Mukherjee A, Walker J, Weyant KB, Schroeder CM (2013) Characterization of flavin-based fluorescent proteins: an emerging class of fluorescent reporters. PLoS ONE 8:e64753
Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20:87–90
Nguyen AW, Daugherty PS (2005) Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol 23:355–360
Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, Littman DR, Reinecker HC (2005) CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307:254–258
Nikitas G, Deschamps C, Disson O, Niault T, Cossart P, Lecuit M (2011) Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J Exp Med 208:2263–2277
Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ (1996) Crystal structure of the Aequorea victoria green fluorescent protein. Science 273:1392–1395
Patterson GH (2004) A new harvest of fluorescent proteins. Nat Biotechnol 22:1524–1525
Patterson GH, Piston DW, Day RN (2001) Fluorescent protein spectra. J Cell Sci 114:837–838
Pérez-Arellano I, Pérez-Martinez G (2003) Optimization of the green fluorescent protein (GFP) expression from a lactose-inducible promoter in Lactobacillus casei. FEMS Microbiol Lett 222:123–127
Phumkhachorn P, Rattanachaikunsopon P, Khunsook S (2007) Use of the gfp gene in monitoring bacteriocin-producing Lactobacillus plantarum N014, a potential starter culture in nham fermentation. J Food Prot 70:419–424
Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ (1992) Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111:229–233
Reid BG, Flynn GC (1997) Chromophore formation in green fluorescent protein. Biochemistry 36:6786–6791
Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P (2001) Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol 2:361–367
Rizzo MA, Davidson MW, Piston DW (2010) Fluorescent protein tracking and detection. In: Goldman RD, Swedlow JR, Spector DL (eds) Live cell imaging: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York, pp 3–34
Rizzo MA, Springer GH, Granada B, Piston DW (2004) An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol 22:445–449
Rizzuto R, Brini M, De Giorgi F, Rossi R, Hein R, Tsien RY, Pozzan T (1996) Double labeling of subcellular structures with organelle-targeted GFP mutants in vivo. Curr Biol 6:183–188
Rossini V, Zhurina D, Radulovic K, Manta C, Walther P, Riedel CU, Niess JH (2014) CX3CR1+ cells facilitate the activation of CD4 T cells in the colonic lamina propria during antigen-driven colitis. Mucosal Immunol 7:533–548
Ruiz L, Motherway MO, Lanigan N, van Sinderen D (2013) Transposon mutagenesis in Bifidobacterium breve: construction and characterization of a Tn5 transposon mutant library for Bifidobacterium breve UCC2003. PLoS ONE 8:e64699
Rush CM, Hafner LM, Timms P (1994) Genetic modification of a vaginal strain of Lactobacillus fermentum and its maintenance within the reproductive tract after intravaginal administration. J Med Microbiol 41:272–278
Russo P, Iturria I, Mohedano ML, Caggianiello G, Rainieri S, Fiocco D, Pardo MA, López P, Spano G (2015) Zebrafish gut colonization by mCherry-labelled lactic acid bacteria. Appl Microbiol Biotechnol 99:3479–3490
Sarxelin M, Tynkkynen S, Mattila-sandholm T, de Vos WM (2005) Probiotic and other functional microbes: from markets to mechanisms. Curr Opin Microbiol 16:204–211
Scott KP, Mercer DK, Glover LA, Flint HJ (1998) The green fluorescent protein as a visible marker for lactic acid bacteria in complex ecosystems. FEMS Microbiol Ecol 26:219–230
Scott KP, Mercer DK, Richardson AJ, Melville CM, Glover LA, Flint HJ (2000) Chromosomal integration of the green fluorescent protein gene in lactic acid bacteria and the survival of marked strains in human gut simulations. FEMS Microbiol Lett 182:23–27
Shagin DA, Barsova EV, Yanushevich YG, Fradkov AF, Lukyanov KA, Labas YA, Semenova TN, Ugalde JA, Meyers A, Nunez JM, Widder EA, Lukyanov SA, Matz MV (2004) GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol Biol Evol 21:841–850
Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572
Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2:905–909
Shaner NC, Patterson GH, Davidson MW (2007) Advances in fluorescent protein technology. J Cell Sci 120:4247–4260
Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY (2008) Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5:545–551
Shcherbo D, Murphy CS, Ermakova GV et al (2009) Far-red fluorescent tags for protein imaging in living tissues. Biochem J 418:567–574
Shimomura O, Johnson FH, Saiga Y (1962) Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol 59:223–239
Southward CM, Surette MG (2002) The dynamic microbe: green fluorescent protein brings bacteria to light. Mol Microbiol 45:1191–1196
Stewart PS, Franklin MJ (2008) Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210
Sugimura Y, Hagi T, Hoshino T (2011) Correlation between in vitro mucus adhesion and the in vivo colonization ability of lactic acid bacteria: screening of new candidate carp probiotics. Biosci Biotechnol Biochem 75:511–515
Sullivan KF (1999) Enlightening mitosis: construction and expression of green fluorescent fusion proteins. Methods Cell Biol 61:113–135
Sun Z, Baur A, Zhurina D, Yuan J, Riedel CU (2012) Accessing the inaccessible: molecular tools for bifidobacteria. Appl Environ Microbiol 78:5035–5042
Tauer C, Heinl S, Egger E, Heiss S, Grabher R (2014) Tuning constitutive recombinant gene expression in Lactobacillus plantarum. Microb Cell Fact 13:150
Toda T, Kosaka H, Terai M, Mori H, Benno Y, Yamori Y (2005) Effects of fermented milk with Lactococcus lactis subsp cremoris FC on defecation frequency and fecal microflora in healthy elderly volunteers. J Jpn Soc Food Sci Technol 52:243–250
Tsien R (1998) The green fluorescent protein. Ann Rev Biochem 67:509–544
van Zyl WF, Deane SM, Dicks LM (2015a) Reporter systems for in vivo tracking of lactic acid bacteria in animal model studies. Gut Microbes 6:291–299
van Zyl WF, Deane SM, Dicks LM (2015b) Use of the mCherry fluorescent protein to study intestinal colonization by Enterococcus mundtii ST4SA and Lactobacillus plantarum 423 in mice. Appl Environ Microbiol 81:5993–6002
Verkhusha VV, Lukyanov KA (2004) The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat Biotechnol 22:289–298
Wachter RM, Elsliger MA, Kallio K, Hanson GT, Remington SJ (1998) Structural basis of spectral shifts in the yellow-emission variants of green fluorescent protein. Structure 6:1267–1277
Wang L, Jackson WC, Steinbach PA, Tsien RY (2004) Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci USA 101:16745–16749
Wang YP, Wang JR, Dai WL (2011) Use of GFP to trace the colonization of Lactococcus lactis WH-C1 in the gastrointestinal tract of mice. J Microbiol Methods 86:390–392
Ward WW (2006) Biochemical and physical properties of green fluorescent protein. In: Chalfie M, Kain SR (eds) Green fluorescent protein: properties, applications, and protocols. Wiley-Interscience, New York, pp 39–65
Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP (2001) Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864
Wingen M, Potzkei J, Endres S, Casini G, Rupprecht C, Krauss U, Jaeger K, Drepper T, Gensch T (2014) The photophysics of LOV-based fluorescent proteins—new tools for cell biology. Photochem Photobiol Sci 13:875–883
Yu Q, Dong S, Zhu W, Yang Q (2007) Use of green fluorescent protein to monitor Lactobacillus in the gastro-intestinal tract of chicken. FEMS Microbiol Lett 275:207–213
Zacharias DA, Violin JD, Newton AC, Tsien RY (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913–916
Zapata-Hommer O, Griesbeck O (2003) Efficiently folding and circular permuted variants of the Sapphire mutant of GFP. BMC Biotechnol 3:5–11
Zhang J, Campbell RE, Ting AY, Tsien RY (2002) Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol 3:906–918
Zhang C, Xing XH, Lou K (2005) Rapid detection of a gfp-marked Enterobacter aerogenes under anaerobic conditions by aerobic fluorescence recovery. FEMS Microbiol Lett 249:211–218
Acknowledgments
This work was supported by project RTA2013-00029-00-00 from the Spanish Ministry of Economy and Competitiveness (MINECO). J.M. Landete has a postdoctoral contract with the research program “Ramón y Cajal” from MINECO.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Landete, J.M., Medina, M. & Arqués, J.L. Fluorescent reporter systems for tracking probiotic lactic acid bacteria and bifidobacteria. World J Microbiol Biotechnol 32, 119 (2016). https://doi.org/10.1007/s11274-016-2077-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-016-2077-5