Abstract
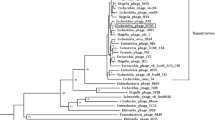
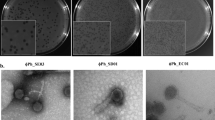
Multiple pathogenic types or serotypes restrict treatment for colibacillosis. In addition, rising antibiotic resistance has heightened public awareness to prevent and control pathogenic Escherichia coli. The bacteriophage is a viable technique to treat colibacillosis as an alternative to antibiotics. P762, a coliphage isolated from duck farm sewage, was demonstrated to cloud lyse Shiga toxin-producing Escherichia Coli serotypes O157 and non-O157 (17/39), Avian pathogenic E. coli covered serotype O78, O83, and O9 (5/19), and other pathogenic Escherichia coli (5/17). Additional fundamental biological characteristics analysis revealed that P762 is stable at pH 3 ~ 11 and temperature between 4 °C and 60 °C, and its optimum multiplicity of infection (MOI) is 0.1. The one-step curve of P762 exhibited three bursts of growth stage: two rapid and one slow stage. Furthermore, the first rapid burst size is 80 CFU/PFU, the burst size of the slow stage is 10 CFU/PFU, and the second rapid burst size is about 990 CFU/PFU. In addition, P762 can form a "halo" on a double agar plate, implying that the phage secretes depolymerase. With 95.14% identity and 90% query coverage, genome sequence analysis revealed that P762 is most closely related to Escherichia phage DY1, which belongs to the genus Kayfunavirus. After screening using RAST and VFDB, no virulence factors were discovered in P762. In vitro antibacterial tests revealed that P762 has high bactericidal activity in lettuce leaves contaminated with STEC. In conclusion, phage P762 might be employed in the future to prevent and control pathogenic Escherichia coli.









Similar content being viewed by others
Data availability
The annotated whole-genome sequence of phage P762 were deposited in the GenBank database under accession number: MW876471.
References
Ron EZ (2006) Host specificity of septicemic Escherichia coli: human and avian pathogens Eliora Z Ron. Curr Opin Microbiol 9:28–32. https://doi.org/10.1016/j.mib.2005.12.001
Croxen MA, Finlay BB (2010) Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 8(1):26–38. https://doi.org/10.1038/nrmicro2265
Wells JG, Davis BR, Wachsmuth IK et al (1983) Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J Clin Microbiol 18(3):512–520. https://doi.org/10.1128/JCM.18.3.512-520.1983
Brooks JT, Sowers EG, Wells JG et al (2005) Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis 192(8):1422–1429. https://doi.org/10.1086/466536
Moxley RA (2004) Escherichia coli O157:H7: an update on intestinal colonization and virulence mechanisms. Anim Health Res Rev 5(1):15–33. https://doi.org/10.1079/AHRR200463
Caprioli A, Morabito S, Brug Re H et al (2005) Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet Res 36(3):289–311. https://doi.org/10.1051/vetres:2005002
Atnafie B, Paulos D, Abera M et al (2017) Occurrence of Escherichia coli O157:H7 in cattle feces and contamination of carcass and various contact surfaces in abattoir and butcher shops of Hawassa Ethiopia. BMC Microbiol 17(1):24. https://doi.org/10.1186/s12866-017-0938-1
Cox GJM, Griffith B, Reed M et al (2020) A vaccine to prevent egg layer peritonitis in chickens. Avian Dis 65(1):198–204. https://doi.org/10.1637/aviandiseases-D-20-00093
Lima Barbieri N, Nielsen DW, Wannemuehler Y et al (2017) mcr-1 identified in avian pathogenic Escherichia coli (APEC). PLoS ONE 12(3):e172997. https://doi.org/10.1371/journal.pone.0172997
Solà-Ginés M, Cameron-Veas K, Badiola I et al (2015) Diversity of multi-drug resistant Avian Pathogenic Escherichia coli (APEC) causing outbreaks of colibacillosis in broilers during 2012 in Spain. PLoS ONE 10(11):e143191. https://doi.org/10.1371/journal.pone.0143191
Smith KE, Wilker PR, Reiter PL et al (2012) Antibiotic treatment of Escherichia coli O157 infection and the risk of hemolytic uremic syndrome. Minnesota Pediatr Infect Dis J 31(1):37–41. https://doi.org/10.1097/INF.0b013e31823096a8
Ramstad SN, Taxt AM, Naseer U et al (2021) Effects of antimicrobials on Shiga toxin production in high-virulent Shiga toxin-producing Escherichia coli. Microb Pathogenesis 152:104636. https://doi.org/10.1016/j.micpath.2020.104636
D’Herelle F (2007) On an invisible microbe antagonistic toward dysenteric bacilli: brief note by Mr. F. D ’ Herelle, presented by Mr. Roux. Res Microbiol 158(7):553–554. https://doi.org/10.1016/j.resmic.2007.07.005
Salmond GPC, Fineran PC (2015) A century of the phage: past, present and future. Nat Rev Microbiol 13(12):777–786. https://doi.org/10.1038/nrmicro3564
Clokie MR, Millard AD, Letarov AV et al (2011) Phages in nature. Bacteriophage 1(1):31–45. https://doi.org/10.4161/bact.1.1.14942
Coffey B, Rivas L, Duffy G et al (2011) Assessment of Escherichia coli O157:H7-specific bacteriophages e11/2 and e4/1c in model broth and hide environments. Int J Food Microbiol 147(3):188–194. https://doi.org/10.1016/j.ijfoodmicro.2011.04.001
Xu J, Chen M, He L et al (2016) Isolation and characterization of a T4-like phage with a relatively wide host range within Escherichia coli. J Basic Microbiol 56(4):405–421. https://doi.org/10.1002/jobm.201500440
Oliveira A, Sereno R, Azeredo J (2010) In vivo efficiency evaluation of a phage cocktail in controlling severe colibacillosis in confined conditions and experimental poultry houses. Vet Microbiol 146(3–4):303–308. https://doi.org/10.1016/j.vetmic.2010.05.015
Badawy S, Baka ZAM, Abou-Dobara MI et al (2022) Biological and molecular characterization of fEg-Eco19, a lytic bacteriophage active against an antibiotic-resistant clinical Escherichia coli isolate. Arch Virol. https://doi.org/10.1007/s00705-022-05426-6
Li M, Guo M, Chen L et al (2020) Isolation and characterization of novel lytic bacteriophages infecting epidemic carbapenem-resistant Klebsiella pneumoniae strains. Front Microbiol. https://doi.org/10.3389/fmicb.2020.01554
Chen M, Zhang L, Abdelgader SA et al (2017) Alterations in gp37 expand the host range of a T4-like phage. Appl Environ Microbiol. https://doi.org/10.1128/AEM.01576-17
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680–685. https://doi.org/10.1038/227680a0
Pajunen M, Kiljunen S, Skurnik M (2000) Bacteriophage φYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to Coliphages T3 and T7. J Bacteriol 182(18):5114–5120. https://doi.org/10.1128/JB.182.18.5114-5120.2000
Ji Y, Cheng M, Zhai S et al (2019) Preventive effect of the phage VB-SavM-JYL01 on rabbit necrotizing pneumonia caused by Staphylococcus aureus. Vet Microbiol 229:72–80. https://doi.org/10.1016/j.vetmic.2018.12.021
Bankevich A, Nurk S, Antipov D et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Aziz RK, Bartels D, Best AA et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. https://doi.org/10.1186/1471-2164-9-75
Grant JR, Stothard P (2008) The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. https://doi.org/10.1093/nar/gkn179
Liu B, Zheng D, Jin Q et al (2019) VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47(D1):D687–D692. https://doi.org/10.1093/nar/gky1080
Kelley LA, Mezulis S, Yates CM et al (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858. https://doi.org/10.1038/nprot.2015.053
Ren J, Wen L, Gao X et al (2009) DOG 1.0: illustrator of protein domain structures. Cell Res 19(2):271–273. https://doi.org/10.1038/cr.2009.6
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Page AJ, Cummins CA, Hunt M et al (2015) Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31(22):3691–3693. https://doi.org/10.1093/bioinformatics/btv421
Xu S, Campisi E, Li J et al (2021) Decontamination of Escherichia coli O157:H7 on fresh Romaine lettuce using a novel bacteriophage lysin. Int J Food Microbiol 341:109068. https://doi.org/10.1016/j.ijfoodmicro.2021.109068
Huang C, Shi J, Ma W et al (2018) Isolation, characterization, and application of a novel specific Salmonella bacteriophage in different food matrices. Food Res Int 111:631–641. https://doi.org/10.1016/j.foodres.2018.05.071
Vengarai Jagannathan B, Kitchens S, Priyesh Vijayakumar P et al (2021) Efficacy of bacteriophage cocktail to control E. coli O157:H7 contamination on baby spinach leaves in the presence or absence of organic load. Microorganisms 9(3):544. https://doi.org/10.3390/microorganisms9030544
Niu YD, Stanford K, Kropinski AM, Ackermann HW, Johnson RP, She YM, Ahmed R, Villegas A, McAllister TA (2012) Genomic, Proteomic and Physiological Characterization of a T5-like Bacteriophage for Control of Shiga Toxin- Producing Escherichia coli O157:H7. PLoS ONE 4(7):e34585
Pereira C, Silva YJ, Santos AL et al (2011) Bacteriophages with potential for inactivation of fish pathogenic bacteria: survival, host specificity and effect on bacterial community structure. Mar Drugs 9(11):2236–2255. https://doi.org/10.3390/md9112236
Salem M, Skurnik M (2018) Genomic characterization of sixteen yersinia enterocolitica-infecting podoviruses of pig origin. Viruses 10(4):174. https://doi.org/10.3390/v10040174
Leungtongkam U, Thummeepak R, Kitti T et al (2020) Genomic analysis reveals high virulence and antibiotic resistance amongst phage susceptible Acinetobacter baumannii. Sci Rep. https://doi.org/10.1038/s41598-020-73123-y
Jayaprakash AD, Jabado O, Brown BD et al (2011) Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res 39(21):e141. https://doi.org/10.1093/nar/gkr693
Hamdi S, Rousseau GM, Labrie SJ et al (2017) Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci Rep. https://doi.org/10.1038/srep40349
Hong Nhung P, Ohkusu K, Mishima N et al (2007) Phylogeny and species identification of the family Enterobacteriaceae based on dnaJ sequences. Diagn Micr Infec Dis 58(2):153–161. https://doi.org/10.1016/j.diagmicrobio.2006.12.019
Delmas J, Breysse F, Devulder G et al (2006) Rapid identification of Enterobacteriaceae by sequencing DNA gyrase subunit B encoding gene. Diagn Micr Infec Dis 55(4):263–268. https://doi.org/10.1016/j.diagmicrobio.2006.02.003
Filippov AA, Sergueev KV, He Y, Huang XZ, Gnade BT, Mueller AJ, Fernandez-Prada CM, Nikolich MP (2012) Bacteriophage-resistant mutants in Yersiniapestis: identification of phage receptors and attenuation for mice. Adv Exp Med Biol 337(954):48. https://doi.org/10.1371/journal.pone.0025486
Hammad AMM (1998) Evaluation of alginate-encapsulated Azotobacter chroococcum as a phage-resistant and an effective inoculum. J Basic Microb 38(1):9–16. https://doi.org/10.1002/(SICI)1521-4028(199803)38:1%3c9::AID-JOBM9%3e3.0.CO;2-4
Mi L, Liu Y, Wang C et al (2019) Identification of a lytic Pseudomonas aeruginosa phage depolymerase and its anti-biofilm effect and bactericidal contribution to serum. Virus Genes 55(3):394–405. https://doi.org/10.1007/s11262-019-01660-4
Li M, Li P, Chen L et al (2021) Identification of a phage-derived depolymerase specific for KL64 capsule of Klebsiella pneumoniae and its anti-biofilm effect. Virus Genes. https://doi.org/10.1007/s11262-021-01847-8
Seul A, Müller JJ, Andres D et al (2014) Bacteriophage P22 tailspike: structure of the complete protein and function of the interdomain linker. Acta Crystallogr D Biol Crystallogr 70(5):1336–1345. https://doi.org/10.1107/S1399004714002685
Barbirz S, Müller JJ, Uetrecht C et al (2008) Crystal structure ofEscherichia coliphage HK620 tailspike: podoviral tailspike endoglycosidase modules are evolutionarily related. Mol Microbiol 69(2):303–316. https://doi.org/10.1111/j.1365-2958.2008.06311.x
Pertics BZ, Cox A, Nyúl A et al (2021) Isolation and characterization of a novel lytic bacteriophage against the K2 capsule-expressing hypervirulent klebsiella pneumoniae strain 52145, and Identification of its functional depolymerase. Microorganisms 9(3):650. https://doi.org/10.3390/microorganisms9030650
Plattner M, Shneider MM, Arbatsky NP et al (2019) Structure and function of the branched receptor-binding complex of bacteriophage CBA120. J Mol Biol 431(19):3718–3739. https://doi.org/10.1016/j.jmb.2019.07.022
Benjamin MMUS, Datta AR (1995) Acid tolerance of enterohemorrhagic Escherichia coli. Appl Environ Microb 61(4):1669–1672. https://doi.org/10.1128/AEM.61.4.1669-1672.1995
Brackett RE, Hao YY, Doyle MP (1994) Ineffectiveness of hot acid sprays to decontaminate Escherichia coli 0157:H7 on Beef. J Food Protect 57(3):198–203. https://doi.org/10.4315/0362-028X-57.3.198
Thomas DE, Elliott EJ (2013) Interventions for preventing diarrhea-associated hemolytic uremic syndrome: systematic review. BMC Public Health 13:799. https://doi.org/10.1186/1471-2458-13-799
Funding
This study was supported by by the following funds: Xinjiang Joint Project of National Natural Science Foundation of China: Targeted screening of virulent phage and its prevention and control in E.coli.( U1803109). Modern agriculture (waterfowl) industrial technology system innovation team for disease prevention and control of Jiangsu (BE2017654). The project of supporting outstanding young talents in universities of anhui province (gxyq2019201). Wuhu Institute of Technology level science and technology team (wzykjtd202002). Animal epidemic prevention and quarantine teaching team of Anhui quality engineering project (2020jxtd282).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Research involving human and animals participants
This article did not contain any study with human participants or animals.
Additional information
Edited by Andrew Millard.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kong, X., Wang, H., Guo, G. et al. Duck sewage source coliphage P762 can lyse STEC and APEC. Virus Genes 58, 436–447 (2022). https://doi.org/10.1007/s11262-022-01915-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-022-01915-7