Abstract
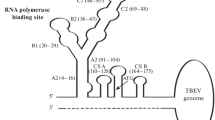
Tick-borne encephalitis viruses (TBEVs) are usually divided into three major subtypes: European (TBEV-Eu), Siberian (TBEV-Sib) and Far Eastern (TBEV-FE). The TBEV-Eu strains have the longest genomes, and TBEV-FE strains have the smallest genomes. Changes in the variable region of the untranslated region (V3′ UTR) play a major role in determining the viral genome length. Analyses of the 3′ UTRs of the different subtypes of TBEV have revealed significant changes in the secondary structures of the V3′ UTR of TBEV. More complex secondary structures of the V3′ UTR regions are typical for TBEV-Eu. The Siberian strain Tomsk-PT122 was isolated from birds and has an unusual 3′ UTR. Several short fragment (24–26 nucleotides) insertions derived from the viral E (2) and NS4a (1) genes have been found in the V3′ UTR of Tomsk-PT122. Additionally, the length of the V3′ UTR increases from 21 to 37 nucleotides during passages of the C11-13 strain of TBEV-Sib into PEK, 293 and Neuro-2a cells. The elongation of the V3′ UTRs of Tomsk-PT122 and C11-13 is the first direct evidence of an intragenomic 3′ UTR modification (insertion) for TBEV. Thus, the obtained results suggest that changing the length of the V3′ UTR in the genome is typical for different TBEV subtypes and can play an essential role in effective TBEV replication in different host cells.




Similar content being viewed by others
References
Grard G, Moureau G, Charrel RN, Lemasson JJ, Gonzalez JP, Gallian P, Gritsun TS, Holmes EC, Gould EA, de Lamballerie X (2007) Genetic characterization of tick-borne flaviviruses: new insights into evolution, pathogenetic determinants and taxonomy. Virology 361:80–92. https://doi.org/10.1016/j.virol.2006.09.015
Gritsun TS, Lashkevich VA, Gould EA (2003) Tick-born encephalitis. Antiviral Res 57:129–146
Gritsun TS, Venugopal K, Zanotto PM, Mikhailov MV, Sall AA, Holmes EC, Polkinghorne I, Frolova TV, Pogodina VV, Lashkevich VA, Gould EA (1997) Complete sequence of two tick-borne flaviviruses isolated from Siberia and the UK: analysis and significance of the 5′ and 3′ UTRs. Virus Res 49:27–39
Proutski V, Gritsun TS, Gould EA, Holmes EC (1999) Biological consequences of deletions within the 3′-untranslated region of flaviviruses may be due to rearrangements of RNA secondary structure. Virus Res 64:107–123
Markoff L (2003) 5′- and 3′-noncoding regions in flavivirus RNA. Adv Virus Res 59:177–228
Gritsun TS, Gould EA (2006) The 3′ untranslated region of tick-borne flaviviruses originated by the duplication of long repeat sequences within the open reading frame. Virology 350:269–275. https://doi.org/10.1016/j.virol.2006.03.002
Thurner C, Witwer C, Hofacker IL, Stadler PF (2004) Conserved RNA secondary structures in Flaviviridae genomes. J Gen Virol 85(Pt 5):1113–1124. https://doi.org/10.1099/vir.0.19462-0
Men R, Bray M, Clark D, Chanock RM, Lai CJ (1996) Dengue type 4 virus mutants containing deletions in the 3’ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J Virol 70:3930–3937
Mandl CW, Aberle JH, Aberle SW, Holzmann H, Allison SL, Heinz FX (1998) In vitro-synthesized infectious RNA as an attenuated live vaccine in a flavivirus model. Nat Med 4(12):1438–1440. https://doi.org/10.1038/4031
Gritsun TS, Gould EA (2006) Direct repeats in the 3’ untranslated regions of mosquito-borne flaviviruses: possible implications for virus transmission. J Gen Virol 87(Pt 11):3297–3305. https://doi.org/10.1099/vir.0.82235-0
Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH (1987) Conserved elements in the 3’ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol 198:33–41
Charlier N, Leyssen P, Pleij CW, Lemey P, Billoir F, Van Laethem K, Vandamme AM, De Clercq E, de Lamballerie X, Neyts J (2002) Complete genome sequence of Montana Myotis leukoencephalitis virus, phylogenetic analysis and comparative study of the 3’ untranslated region of flaviviruses with no known vector. J Gen Virol 83(Pt 8):1875–1885. https://doi.org/10.1099/0022-1317-83-8-1875
Pletnev AG (2001) Infectious cDNA clone of attenuated Langat tick-borne flavivirus (strain E5) and a 3’ deletion mutant constructed from it exhibit decreased neuroinvasiveness in immunodeficient mice. Virology 282:288–300. https://doi.org/10.1006/viro.2001.0846
Chausov EV, Ternovoi VA, Protopopova EV, Kononova JV, Konovalova SN, Pershikova NL, Romanenko VN, Ivanova NV, Bolshakova NP, Moskvitina NS, Loktev VB (2010) Variability of the tick-borne encephalitis virus genome in the 5’ noncoding region derived from ticks Ixodes persulcatus and Ixodes pavlovskyi in Western Siberia. Vector Borne Zoonotic Dis 10:365–375. https://doi.org/10.1089/vbz.2009.0064
Ecker M, Allison SL, Meixner T, Heinz FX (1999) Sequence analysis and genetic classification of tick-born encephalitis viruses from Europe and Asia. J Gen Virol 80:179–185. https://doi.org/10.1099/0022-1317-80-1-179
Ternovoi VA, Protopopova EV, Chausov EV, Novikov DV, Leonova GN, Netesov SV, Loktev VB (2007) Novel variant of tickborne encephalitis virus, Russia. Emerg Infect Dis 13:1574–1578. https://doi.org/10.3201/eid1310.070158
Gritsun DJ, Jones IM, Gould EA, Gritsun TS (2014) Molecular archaeology of Flaviviridae untranslated regions: duplicated RNA structures in the replication enhancer of Flaviviruses and Pestiviruses emerged via convergent evolution. PLoS ONE. https://doi.org/10.1371/journal.pone.0092056
Wallner G, Mandl CW, Kunz C, Heinz FX (1995) The flavivirus 3’-noncoding region: extensive size heterogeneity independent of evolutionary relationships among strains of tick-borne encephalitis virus. Virology 213:169–178. https://doi.org/10.1006/viro.1995.1557
Sakai M, Yoshii K, Sunden Y, Yokozawa K, Hirano M, Kariwa H (2014) Variable region of the 3’ UTR is a critical virulence factor in the Far-Eastern subtype of tick-borne encephalitis virus in a mouse model. J Gen Virol 95(Pt 4):823–835. https://doi.org/10.1099/vir.0.060046-0
Sakai M, Muto M, Hirano M, Kariwa H, Yoshii K (2015) Virulence of tick-borne encephalitis virus is associated with intact conformational viral RNA structures in the variable region of the 3’-UTR. Virus Res 203:36–40. https://doi.org/10.1016/j.virusres.2015.03.006
Muto M, Kamitani W, Sakai M, Hirano M, Kobayashi S, Kariwa H, Yoshii K (2018) Identification and analysis of host proteins that interact with the 3’-untranslated region of tick-borne encephalitis virus genomic RNA. Virus Res 249:52–56. https://doi.org/10.1016/j.virusres.2018.03.006
Ponomareva EP, Ternovoi VA, Mikryukova TP, Protopopova EV, Gladysheva AV, Shvalov AN, Konovalova SN, Chausov EV, Loktev VB (2017) Adaptation of tick-borne encephalitis virus from human brain to different cell cultures induces multiple genomic substitutions. Arch Virol 162:3151–3156. https://doi.org/10.1007/s00705-017-3442-x
Chausov EV, Ternovoy VA, Protopopova EV, Konovalova SN, Kononova Yu V, Tupota NL, Moskvitina NS, Romanenko VN, Ivanova NV, Bol’shakova NP, Leonova GN, Loktev VB (2011) Molecular genetic analysis of the complete genome of tick-borne encephalitis virus (Siberian subtype): modern Kolarovo-2008 isolate. Probl Part Danger Infect 110:44–48 (in Russian)
Demina TV, Dzhioev IuP, Kozlova IV, Verkhozina MM, Tkachev SE, Doroshchenko EK, Lisak OV, Paramonov AI, Zlobin VI (2012) Genotypes 4 and 5 of the tick-borne encephalitis virus: features of the genome structure and possible scenario for its formation. Vopr Virusol 57(4):13–19 (in Russian)
Dai X, Shang G, Lu S, Yang J, Xu J (2018) A new subtype of eastern tick-borne encephalitis virus discovered in Qinghai-Tibet Plateau, China. Emerg Microbes Infect 7(1):74. https://doi.org/10.1038/s41426-018-0081-6
Bertrand YJ, Johansson M, Norberg P (2016) Revisiting recombination signal in the tick-borne encephalitis virus: a simulation approach. PLoS ONE. https://doi.org/10.1371/journal.pone.0164435
Dzhioev YP, Paramonov A, Reva ON, Bukin YS, Kozlova IV, Demina TV, Tkachev SE, Zlobin VI (2015) Detection of potential sites of recombination in the tick-borne encephalitis virus by the methods of comparative genomics. Vopr Virusol 60(3):44–49 (in Russian)
Acknowledgements
The authors would like to gratefully acknowledge support from the Russian Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing (GZ for SRC VB “Vector” № 141-00069-18-01 from 17.03.2018).
Author information
Authors and Affiliations
Contributions
VAT and VBL conceived and designed the experiments and wrote the manuscript. AVG and ANS performed the experiments and analysed the data and assisted in writing the manuscript. EPP performed the experiments and assisted in the writing of manuscript. TMP, EVA, SNK and EVC performed the experiments.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest. No competing financial interests exist.
Research involving human participants and/or animals
The present study did not involve any human subject or animal.
Informed consent
The present study did not involve any human subject and therefore, no need of informed consent.
Additional information
Edited by Joachim Jakob Bugert.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ternovoi, V.A., Gladysheva, A.V., Ponomareva, E.P. et al. Variability in the 3′ untranslated regions of the genomes of the different tick-borne encephalitis virus subtypes. Virus Genes 55, 448–457 (2019). https://doi.org/10.1007/s11262-019-01672-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-019-01672-0