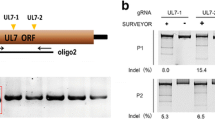
Abstract
The UL24 homologous genes are conserved in alphaherpesviruses. However, the proximity of the UL24 gene and the UL23 gene encoding for thymidine kinase (TK) in the genome of suid herpesvirus 1 (SuHV-1) makes it difficult to mutate UL24 without affecting the expression of the TK gene, and thus functional studies of the UL24 gene have lagged behind. In this study, CRISPR/Cas9 and homologous recombination were adopted to generate UL24 and TK mutant viruses. Deletion of either the UL24 or the TK gene resulted in significantly reduced SuHV-1 replication and spread capacity in Vero cells. However, UL24-deleted virus still maintained a certain degree of lethality in mice, while TK-deleted viruses completely lost their lethality in mice. Similarly, neurovirulence of UL24-deleted virus in mice was not significantly affected compared to parental virus. In comparison, infection with the TK-deleted viruses resulted in significantly reduced neurovirulence and complete loss of lethality. In addition, and for the first time, viral UL24 protein was found to be expressed late during SuHV-1 infection; enhanced green fluorescence protein (eGFP) labeled UL24 protein was shown to be localized in the nucleus via heterologous expression. In conclusion, the UL24 gene of SuHV-1 encodes a nuclear-localized viral protein and acts as a minor virulence-associated factor compared to the TK gene.







Similar content being viewed by others
References
Pellett PE, Davison AJ, Eberle R, Ehlers B, Hayward GS, Lacoste V et al (2011) Herpesvirales. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: Ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press, London, p 99–107
Pejsak ZK, Truszczyni MJ (2006) Truszczynsk: Aujeszky’s disease (pseudorabies). In: Straw BE, Zimmerman JJ, D’Allaire S, Taylor DJ (eds) Diseases of swine, 9th ed. Blackwell Publishing Ltd, Ames, p 419–433
An TQ, Peng JM, Tian ZJ, Zhao HY, Li N, Liu YM et al (2013) Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China. Emerg Infect Dis 19:1749–1755
Luo Y, Li N, Cong X, Wang CH, Du M, Li L et al (2014) Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet Microbiol 174:107–115
Yu X, Zhou Z, Hu D, Zhang Q, Han T, Li X et al (2014) Pathogenic pseudorabies virus, China, 2012. Emerg Infect Dis 20:102–104
Ye C, Zhang QZ, Tian ZJ, Zheng H, Zhao K, Liu F et al (2015) Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: evidence for the existence of two major genotypes. Virology 483:32–43
Klupp BG, Hengartner CJ, Thomas C, Enquist LW, Mettenleiter TC (2004) Complete, annotated sequence of the pseudorabies virus genome. J Virol 78:424–440
Pomeranz LE, Reynolds AE, Hengartner CJ (2005) Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev 69:462–500
Bertrand L, Leiva-Torres GA, Hyjazie H, Pearson A (2010) Conserved residues in the UL24 protein of herpes simplex virus 1 are important for dispersal of the nucleolar protein nucleolin. J Virol 84:109–118
Jacobson JG, Chen SH, Cook WJ, Kramer MF, Coen DM (1998) Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161–169
Jacobson JG, Martin SL, Coen DM (1989) A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J Virol 63:1839–1843
Kasem S, Yu MHH, Yamada S, Kodaira A, Matsumura T, Tsujimura K et al (2010) The ORF37 (UL24) is a neuropathogenicity determinant of equine herpesvirus 1 (EHV-1) in the mouse encephalitis model. Virology 400:259–270
Blakeney S, Kowalski J, Tummolo D, DeStefano J, Cooper D, Guo M et al (2005) Herpes simplex virus type 2 UL24 gene is a virulence determinant in murine and guinea pig disease models. J Virol 79:10498–10506
Mali P, Esvelt KM, Church GM (2013) Cas9 as a versatile tool for engineering biology. Nat Methods 10:957–963
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278
Ebina H, Misawa N, Kanemura Y, Koyanagi Y (2013) Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep 3:2510
Bi Y, Sun L, Gao D, Ding C, Li Z, Li Y et al (2014) High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. PLoS Pathog 10:e1004090
Liang X, Sun L, Yu T, Pan Y, Wang D, Hu X et al (2016) A CRISPR/Cas9 and Cre/Lox system-based express vaccine development strategy against re-emerging Pseudorabies virus. Sci Rep 6:19176
Xu A, Qin C, Lang Y, Wang M, Lin M, Li C et al (2015) A simple and rapid approach to manipulate pseudorabies virus genome by CRISPR/Cas9 system. Biotechnol Lett 37:1265–1272
Tong W, Liu F, Zheng H, Liang C, Zhou YJ, Jiang YF et al (2015) Emergence of a pseudorabies virus variant with increased virulence to piglets. Vet Microbiol 181:236–240
Auer TO, Duroure K, Concordet JP, Del Bene F (2014) CRISPR/Cas9-mediated conversion of eGFP-into Gal4-transgenic lines in zebrafish. Nat Protoc 9:2823–2840
Tang YD, Liu JT, Wang TY, An TQ, Sun MX, Wang SJ et al (2016) Live attenuated pseudorabies virus developed using the CRISPR/Cas9 system. Virus Res 225:33–39
De Clercq E (2005) Antiviral drug discovery and development: where chemistry meets with biomedicine. Antiviral Res 67:56–75
Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH (2015) Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids 4:e264
Acknowledgements
This study was supported by the National Key Research and Development Program of China (2016YFD0500100), China Agriculture Research System (NYCYTX-009), and Shanghai Municipal Agriculture Science and Technology Key Project (2015, 1-7 and 2016, 4-2).
Author information
Authors and Affiliations
Contributions
CY and JC conducted the most experiments and drafted the manuscript. GT critically revised the manuscript. XC, SZ, SJ, JX, HZ, WT, GL helped with the experiments. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no confict of interest.
Ethical approval
All applicable international, national, and institutional guidelines for the care and use of animals were followed.
Research involving human and animal participants
The animal experiments were performed according to the Guide for the Care and Use of Laboratory Animals of Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, China. The animal ethics approved number was SHVRI-MO-2017070588.
Additional information
Communicated by William Dundon.
Rights and permissions
About this article
Cite this article
Ye, C., Chen, J., Cheng, X. et al. Functional analysis of the UL24 protein of suid herpesvirus 1. Virus Genes 55, 76–86 (2019). https://doi.org/10.1007/s11262-018-1619-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-018-1619-3