Abstract
The Orf virus 050 (ORFV050) gene is located in the core region of the ORFV genome. It is similar to Vaccinia virus (VV) Copenhagen L4R, and encodes the DNA-binding virion core protein VP8, which has structures similar to the VV P25K core protein and may undergo similar proteolytic processing during virus assembly. Three conserved Ala–Gly–X motifs at putative cleavage sites were identified in ORFV050. To investigate the proteolysis of ORFV050 and its participation in viral assembly, full-length and site-directed mutant ORFV050 recombinant proteins were constructed and expressed. Two distinct protein bands of 28.5 and 25 kDa were detected in the infected cells using anti-ORFV050 polyclonal antiserum. A potential cleavage site was identified at amino acids 30–32 of ORFV050. Mutation of AG/A to (R) in ORFV050 abolished the process of proteolysis. ORFV050 is a late gene synthesized during viral replication in the host cytoplasm. According to these results, we conclude that ORFV050 undergoes proteolysis and plays an important role in viral assembly.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As an infectious viral zoonosis, ovine-contagious pustular dermatitis (Orf) occurs worldwide and has been reported in many provinces in China. Orf virus (ORFV) typically infects wild ruminant species, such as goats, sheep, and deer, with lambs and immunodeficient individuals being particularly susceptible. Human infection has been reported [1, 2]. ORFV is a dsDNA belonging to parapoxvirus, a prototype member of the family Poxviridae, and replicates within the cytoplasm of the infected host cells. The genome of ORFV is approximately 138 kbp, has a G+C content of 64%, and contains 89 conserved genes (ORF009–ORF111) in a total of 131 putative genes [3, 4].
ORFV050 is a highly conserved gene which is located in the middle of the ORFV genome. The ORFV050 gene of the NA1/11 strain is 780 bp and encodes a 28.5 kDa late protein which is thought to be necessary for proper viral genome packaging and assembly [5]. The proteolysis of core protein in Vaccinia virus (VV) happened at putative cleavage sites with Ala–Gly–X motifs. In addition to the AG/X motif, GG/X motifs also undergo the proteolytic processing in adenovirus, Africa swine fever (ASF) [6] and ORFV [7]. The structural protein ORFV050, located within virion core of ORFV, has three putative cleavage sites in both N-terminus and C-terminus, and may play an important role in viral replication and morphogenesis. Because the molecular weight of the hydrolytic product of the AG/A cleavage site in the C-terminal region (253–255aa) is only 0.5 kDa, and hard to detect, we focus solely on the N-terminal AGF/AGA cleavage sites in this study. Earlier studies [8–10] showed that the proteolytic processing of VV happened at the AGA site. On this basis, we choose the corresponding AGA (30–32aa) site for this research.
As with the VV, the core protein of other DNA viruses, such as the African swine fever virus, requires the cleavage of precursor proteins to complete their infectious life cycle, undergoing an essential mechanism for viral replication and morphogenesis [6]. It is expected that the ORFV may undergo proteolytic cleavage during infection. Several major structural core proteins of VV occurring during the late stages of core formation, such as P4a (A10L), P4b (A3L), are catalyzed by VV-encoded proteinase. The proteolytic processing of these core proteins is essential for maturation of the VV progeny. Recent research [7] has proven that ORFV086, which shares a similar structure to the VV P4a core protein, undergoes proteolytic processing. As one of the core proteins of the ORFV, we suspect that ORFV050 may be processed via a similar mechanism, with proteolytic processing occurring at the putative cleavage sites with AG/X motifs.
To confirm our hypothesis, and to verify that proteolytic processing occurs in the predicted AG/X motif sites, full-length ORFV050 and site-directed mutants were constructed and examined. Rifampicin was chosen to block the proteolytic processing to evaluation its effect on proteolysis.
Materials and methods
Cells and virus strain
As a host of ORFV, primary ovine fetal turbinate (OFTu) cells were cultured in minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), containing l-glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 μg/ml) in a 37 °C incubator with 5% CO2. The NA1/11 strain is a native ORFV strain isolated from a sheep diagnosed with orf in the Jilin province of China. NA1/11 is used as a infection medium of OFTu cells. The concentration of FBS in MEM was reduced to 2% during the subsequent viral infection.
Sequence alignment of ORFV050
To analyze the homology of ORFV050 with different ORFV strains and VV or Cowpox virus, amino acid sequences of ORFV050 and P25K were obtained from Genbank. Sequence alignments of VV Copenhagen P25K protein with ORFV050 proteins among several ORFV strains (NZ2, ABA00567.1; OV-SA00, NP_957827.1; OV-IA82, AAR98145.1; B029, AHH34239.1; D1701, ADY76891.1; NA1/11, and AHZ33747.1) were performed using CLUSTAL W. Accession numbers and other information of the aligned proteins are shown in Table 1.
Plasmid
To construct a prokaryotic expression vector of the ORFV050 protein, the fully sequenced genome of ORFV strain NA1/11 was acquired from Genbank. (Accession No: AHZ33747.1). The coding sequence of ORFV050 was PCR amplified using the primers 32a-050-HisFw (BamHI):5′ GCGGATCCATGACCAATCTGCTTTCGTT 3′ and 32a-050-His Rv (HindIII′): 5′TATAAGCTTACTACGGCGCGGCCTCGGC 3′. The PCR product was subcloned into the expression vector pET32a(+) vector via BamHI/HindIII restriction sites to generate pET32a-ORFV050. In order to express the ORFV050 protein in mammalian cells, its sequence was similarly amplified from the NA1/11 genome utilizing the primers described in Table 2. Amplicon digested by the restriction enzyme was cloned in-frame with a FLAG-tag in the N-terminal of pCMVtag2B vector to generate pCMVtag2B-050, with a FLAG-tag in the C-terminal of pCDNA3.0 vector to generate pCNDA3.0-050 and with a GFP-tag in the pEGFP-N1 vector to generate pEGFP-N1-050. All PCR products were purified using a Cycle Pure Kit, according to the manufacturer’s protocol, and ligated into the pCMV-Tag2B plasmid and pCDNA3.0. Afterward, the quality of all ORFV050 constructs was verified by sequencing.
Expression of ORFV050 protein and production of anti-ORFV050 polyclonal anti-serum
In order to produce a large quantity of recombinant ORFV050 protein (rORFV050) with a His-tag, we constructed the prokaryotic expression plasmid pET32a-050 and expressed it in Escherichia coli BL21 (DH3). The pET32a-050 plasmid was transformed into E. coli BL21 and induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 18 h at 20 °C. The target protein was expressed in both the soluble fraction and inclusion body, and the soluble fraction was purified using a Ni2+-NTA column. Each purified fraction was analyzed using SDS-polyacrylamide (SDS-PAGE) stained with Coomassie Brilliant Blue.
The purified pET32a-050 protein was used as an antigen to produce polyclonal anti-serum. The ORFV050 protein concentration was measured using a Quick Start™ Bradford Protein Assay Kit, and the final concentration obtained was 1.25 mg/ml. Purified protein was mixed with adjuvants in a ratio of 1:1 (v/v) and emulsified by syringe. The mixtures were injected into two 8-week-old male New Zealand rabbits in a low-dose multipoint injection method with the protein mixtures. 200 μg of ORFV050 was emulsified with Freund’s complete adjuvant for primary immunization, followed by emulsification with Freund’s incomplete adjuvant for all subsequent immunizations. About 1 week after the third immunization, small amounts of blood were sampled from the immunized rabbits in order to test titers using Elisa. The polyclonal anti-sera were collected when the titer ratio was 1:80,000.
Transcription kinetics of ORFV050 processing
Kinetics of ORFV050 were investigated during ORFV replication in OFTu cells by reverse transcription-PCR (RT-PCR). OFTu cells were incubated with NA1/11 at MOI (multiplicity of infection) of 5 PFU/cell, both in the presence and absence of cytosine arabinoside (AraC; 40 μg/ml; Sigma, St. Louis, MO, USA), after 16 h of culture. As an inhibitor of DNA replication, AraC mainly affects the late transcription of poxviruses. The infected cells were harvested at 0, 1, 2, 3, 4, 6, 8, 10, 12, and 24 h post-infection. cDNA templates were reverse transcribed from the extracted RNA. The transcriptions of ORFV050 and ORFV119 (early late control gene) were performed by PCR using the primers ORFV050-Flag Fw5′ AAGGTACCATGACCAATCTGCTTTCGTT 3′, ORFV050-Flag Rv5′ GCGATATCCTACTTATCGTCGTCATCCTT 3′, ORFV119 Fw 5′ CCCAAGCTTATGGACTCTCGTAGGC 3′, and ORFV119 Rv 5′ CCGCTCGAGATCGCTGTCGCTGTCG 3′. Primers were all designed based on the NA1/11 sequence (Accession Number: AHZ33747.1). To further analyze the kinetics of ORFV050 protein expression using western blots, infected OFTu cells were collected at different times (0, 3, 5, 8, 10, 12, and 24 h p. i.). All samples were subjected to western blot analysis using anti-ORFV050 polyclonal antiserum.
Construction and site-directed mutation of expression plasmids
In order to identify the cleavage sites of the ORFV050 protein, we constructed plasmids with site-directed mutation at the putative AG/X cleavage sites. Three putative cleavage sites were found in the ORFV050 protein, and the AGA motif at 30–32aa corresponds to the site identified in VV P25K. Site-directed mutation of the putative cleavage site AGA motif of pcDNA3.0-050 was performed using the Muta-direct™ Site-directed Mutagenesis Kit to convert AGA into IDI. Primers of Directed Mutagenesis were designed as follows: 050-AGA-IDI FW 5′GACCATGCTCGTGATCATCGACATGCGAGTGCCTTCCCAC3′, 050-AGA-IDI RV 5′GTGGGAAGCGCACTCGCATGTCGATGATCACGAGCATGGTC3′. The mutation site is shadowed above. The integrity of the constructed plasmid and mutants were verified by sequencing.
Transient expression
80% confluent monolayers of OFTu cells cultured in 6-well plates for 16 h were transfected with 2.5 μg of plasmid using Lipofectamin® 2000, according to the manufacturer’s protocol, following infection or lack of infection with ORFV NA1/11. For each well, 5 μl Lipofectamin was diluted with 100 μl Opti-MEM and incubated for 10 min at room temperature. 2.5 μg of plasmid DNA was diluted with 100 μl Opti-MEM, vortexed, and mixed with liposome at a 1:1 ratio. The DNA–lipid mixture was then incubated for 20 min at room temperature. After incubation, the DNA mixture was added to each well of cells, which had been maintained in 800 μl Opti-MEM, and incubated for 6 h at 37 °C. After the 6 h transfection, the transfection medium was replaced with fresh MEM complemented with 10% FBS, and the cells continued to culture for 2 days.
Western blotting
OFTu cells were transfected with pCDNA3.0-050-FLAG or pCMV-Tag2B-050 and cultured in 6-well plates for 16 h with or without NA1/11. After 48 h of incubation, infected cells were collected, washed in PBS (pH 7.4), and subsequently lysed. Cell lysates were heated at 100 °C for 10 min in loading buffer and subjected to 12% SDS-PAGE analysis. The separated proteins were electrotransferred to PVDF membranes at 120 V for 80 min at 4 °C. The membranes were blocked with 5% skim milk for 2 h. The membranes were respectively probed with anti-ORFV050 polyclonal antiserum and anti-FLAG antibody diluted with TBS, and incubated at room temperature for 2 h. The membranes were washed in TBST (0.1% Tween-20 in TBS) three times, and hybridized with a horseradish peroxidase-conjugated goat anti-mouse and goat anti-rabbit antibody (1:10,000 dilution), respectively. The chemiluminescent substrate (ECL) was added to develop the immunoblots and to observe the ORFV050 protein.
Confocal microscopy
Monolayers of OFTu cells were cultured on plates and cotransfected with pEGFP-N1 (GFP control) or 050EGFP/pEGFP-N1 (ORFV050-EGFP fusion). At 24 h post transfection, the cells were fixed, stained with DAPI and visualized under confocal microscopy (LSM710; Zeiss).
Effect of rifampicin in ORFV050 proteolytic processing
OFTu cells were infected with NA1/11 at MOI = 10 and treated immediately with different concentrations of rifampicin The cells were harvested 24 h post infection. Cell lysates were subjected to SDS-PAGE and electrotransferred to PVDF membranes. The ORFV050 protein was detected using anti-ORFV050 polyclonal antiserum. The densitometric analysis of the blots was performed using ImageJ software, version 1.62 (National Institutes of Health, Bethesda, MD). The statistical analysis of the data was performed by Student’s t test.
Results
ORFV050 protein sequence alignment
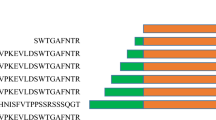
Background data regarding the ORFV050 protein is limited, with only a small number of reference strains having been sequenced. Thus, amino acid sequences of six different ORFV strains and the VV P25K strain were chosen for our comparison. Sequence alignment of different ORFV strains revealed that NA1/11 ORFV050 shares 99.1% identity with the other 5 ORFV strains (Fig. 1a) and 47% similarity with VV Copenhagen strain [11, 12]. Three instances of the AG/X motif were found in each of the 6 ORFV strains at corresponding positions except for a TGA motif at the C-terminal position for strain NZ2. A comparison of the sequences of ORFV050 and VV indicated that the proteolytic sites of P25K at AGS and AGA also exist at the sites of AGF(17–19) and AGA(30–32) of ORFV050, but the ORFV050 AGA(253–255aa) site was not found in P25K VV (Fig. 1b). These results suggest that the AG/X motif is highly conserved in poxviruses, and that ORFV050 may undergo proteolytic processing similar to VV P25K.
Comparison of the amino acid sequences of ORFV050 among different ORFV strains (a) and VV P25K (b). a Amino acid sequences of ORFV050 protein from NA1/11 (AHZ33747.1), OV-IA82 (AAR98145.1), NZ2 (ABA00567.1), D1701 (ADY76891.1), OV-SA00 (NP_957827.1), and B029 (AHH34239.1) were aligned using CLUSTAL W (San Diego Supercomputer Center biology workbench; http://workbench.sdsc.edu/). Nine amino acid mutations among these strains are indicated below the alignment (asterisk). Putative proteolytic processing sites are boxed. b The alignment of ORFV050 and VV (AAA48079.1) was performed using CLUSTAL W. Putative proteolytic processing sites are boxed (Color figure online)
Purification of the ORFV050 protein
As the ORFV strain NA1/11 was discovered in Nongan of Jilin province of China, NA1/11 ORFV050 protein was chosen as the antigen for polyclonal antibody production. In order to obtain large quantities of the NA1/11 ORFV (rORFV050) protein, it was expressed in E. coli transformed with pET32a-ORFV050 (Fig. 2a), and the soluble protein was purified using a nickel column. The predicted molecular weight of ORFV050 protein with His-tag was 47 kDa, which is in agreement with the protein observed in the SDS-PAGE analysis. As shown in the gel stained with Coomassie Brilliant Blue (Fig. 2b), the high purity of ORFV050 protein was enough for it to be used as an immunogen to produce polyclonal antibodies. 3 ml of rORFV050 protein with a concentration of 1.25 mg/ml was purified.
Expression and purification of rORFV050 protein. a ORFV050 protein was prepared from E. coli transformed with pET32a-ORFV050 and then induced by IPTG. E. coli transformed with pET32a was used as control. M marker, S soluble protein, I inclusion body. A 47 kDa protein was found both in S and I. b The soluble protein ORFV050 was applied to a Ni2+-NTA column. Flow-through (F), and the washed fractions were collected sequentially. Each fraction was electrophoresed in SDS-12% polyacrylamide gels and stained with Coomassie Brilliant Blue. The 47 kDa protein shares the same molecular weight as that of rORFV050 protein and is indicated on the right side of the figure. The positions of molecular weight markers are marked on the left side of the figure
Identification of anti-ORFV050 polyclonal antibody
Polyclonal anti-serum was purified by Protein-G affinity chromatography. To characterize the antibodies against ORFV050, SDS-PAGE and immunoblotting were performed. Anti-ORFV050 polyclonal antibodies were mixed with loading buffer and separated via SDS-PAGE. Two distinct bands of heavy chain and light chain were observed, and the density of bands indicates that the purity of polyclonal antibody is about 95%. To assess the ability of the ORFV050 polyclonal anti-serum to recognize natural ORFV050 protein, OFTu cells were infected with NA1/11. Two bands of 28.5 and 25 kDa were detected with molecular weights in accordance with the ORFV050 precursor and its proteolytic fraction, proving that anti-ORFV050 polyclonal anti-serum can detect the natural ORFV050 protein (Fig. 3a). To determine whether or not anti-ORFV050 polyclonal antibody can recognize the protein expressed by mammalian vector, OFTu cells were transfected with pCMVtag2B or pCMVtag2B-050 and then lysed for immunoblotting analysis. No protein bands were detected in the mock OFTu cells or in the cells transfected with pCMV-Tag2B. A band of 28.5 kDa was detected by anti-ORFV050 serum in the cells transfected with pCMVtag2B-050, demonstrating that ORFV050 protein expressed by mammalian vector could be identified by this polyclonal antibody (Fig. 3b). These results indicate that anti-ORFV050 serum can specifically recognize the ORFV050 protein expressed both in naturally occurring virus and in mammalian vectors.
Western blot analysis of the characterization of polyclonal antibody. a OFTu cells were infected with NA1/11 for 24 h and harvested with RIPA Lysis Buffer. Lysates were subjected to SDS-PAGE, and the proteins detected by immunoblotting with anti-ORFV050 polyclonal antibody and GAPDH. b 80% confluent OFTu cells were transfected with pCMVTag2B or pCMVTag2B-ORFV050 plasmids and were lysed 48 h later. The extracts of untreated OFTu cells and cells transfected with pCMVTag2B and pCMVTag2B-ORFV050 were resolved by immunoblot with anti-ORFV050 polyclonal antibody and GAPDH. The markers are shown on the left side of the figure. ORFV050 protein and its hydrolysate are indicated
ORFV050 is a late gene during ORFV infection and located in the cytoplasm
In order to determine whether ORFV050 is an early or late gene during viral replication, RT-PCR and western blotting were performed at a range of times throughout the viral replication cycle. The results showed that transcription of ORFV050 could be detected by RT-PCR starting from 8 h and increasing to 24 h p.i., without treatment with AraC, an inhibitor of poxvirus DNA replication [13, 14]. Conversely, in the presence of AraC, no ORFV050 transcription could be detected (Fig. 4a). In contrast, the synthesis of ORFV119 started at 1 h post infection, both in OFTu cells and AraC-blocked cells, demonstrating that ORFV050 is a late poxviral gene. In the absence of AraC, ORFV050 protein synthesis appears in the western blot analysis (Fig. 4b) beginning at 10 h post infection and continuing until 24 h p.i., while the AraC-treated cells displayed no bands. These results support the results of RT-PCR indicating that ORFV050 is a late gene. In the immunoblotting analysis, the ORFV050 full-length protein and a 25 kDa fragment were observed 24 h post infection, suggesting that these are the ORFV050 protein and its proteolytic fraction, pORFV050. Since the ORFV050 precursor appears 10 h post infection and the pORFV050 appears between 12 and 24 h p.i., this suggests that ORFV050 begins synthesis at the late stage of infection and is then hydrolyzed.
Transcription kinetic analysis of the ORFV050 protein. Transcription kinetics of ORFV050 and ORFV119 (early gene control). a OFTu cells were infected with NA1/11(MOI = 5) followed by treatment or lack of treatment of AraC (40 μg/ml) and harvested at different times as indicated. The RNA was extracted and subjected to RT-PCR depending on the cDNA templates for amplifications of ORFV050 and ORFV119. All amplified products were electrophoresed in 1.0% agarose gels. b OFTu cells were infected with NA1/11 at MOI = 5 with or without AraC treatment. All infected cells were collected at the indicated time and subjected to SDS-PAGE analysis and immunoblotting with antibody against FLAG, ORFV050, and GAPDH. Position of ORFV050 is marked on the right side, and DNA molecular weight standards marked on the left side
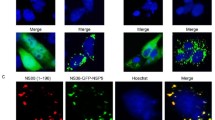
To visualize the subcellular location of the ORFV050 protein, the cells were transfected with pEGPF-N1 or 050EGFP/pEGFP-N1, fixed, stained with DAPI, and visualized under confocal microscopy. The subcellular localization of ORFV050 was investigated in OFTu cells transiently expressing ORFV050-GFP. ORFV050-GFP localized in the cell cytoplasm, as a punctate distribution pattern (Fig. 5).
Subcellular localization of ORFV050. Immunofluorescent assays were performed to identify the subcellular localization of ORFV050 in OFTu cells. OFTu cells were cotransfected with pEGPF-N1 or 050EGFP/pEGFP-N1. 24 h after transfection, cells were fixed with 4% formaldehyde, stained with DAPI, and visualized under confocal microscopy. Green: EGFP (Upper panel); ORFV050GFP (Lower panel). Blue DAPI. BF bright field. Arrows denote cytoplasmic localization. Bar 10 μm (Color figure online)
AGA cleavage site
To further confirm our prediction, the full-length and site-directed mutant vectors were prepared. In order to verify that the cleavage was occurring at the AG/A site (30–32aa), the AG/A (30–32aa) tri-peptide in pCDNA3.0-050 was mutated to IDI, and the mutated vector was transfected into mock-infected and ORFV-infected OFTu cells. Lysates were subjected to immunoblotting analysis with antibody against FLAG and GAPDH.
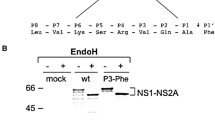
Experiments were divided into two groups: cells treated without ORFV (A group), and cells treated with ORFV (B group) before transfection with pCDNA3.0-050 and pCDNA3.0-IDI-050. In the A group, only a band of 28.5 kDa was observed both in cells transfected with pCDNA3.0-050 and pCDNA3.0-IDI-050. However, in the B group, two bands of 28.5 and 25 kDa were detected in the infected cells transfected with pCDNA3.0-050, but only a 28.5 kDa band appeared in the immunoblot analysis of cells transfected with pCDNA3.0-IDI-050 (Fig. 6).
Immunoblotting analysis of proteolytic cleavage of ORFV050. OFTu cells were transfected with pCDNA3.0-050 or pCDNA3.0 in the presence of absence of NA1/11 (MOI = 5). Transfection was halted by the addition of SDS sample buffer and the proteins detected by western blot, using the antibodies against FLAG and GAPDH. The ORFV050 precursor and its hydrolysate are indicated on the right side of the figure
Rifampicin inhibits the proteolytic processing of ORFV050
As an antibiotic, rifampicin blocks the assembly of VV reversibly by disrupting the viral membrane biogenesis and inhibiting the maturation of the structural core proteins. [15]. To examine the ORFV050 proteolytic processing, rifampicin was used as an inhibitor to block proteolytic processing in ORFV-infected cells. Two distinct bands of 28.5 and 25 kDa were detected in cells infected with NA1/11 but without rifampicin treatment [8, 16], as shown in Fig. 7a. Based on their molecular weights, it was reasonable to assume that they were the ORFV050 precursor and its hydrolysate fraction. As the concentration of rifampicin increased, the syntheses of ORFV050 and its hydrolysate were both reduced, but the reduction of hydrolysate was obviously greater than that of ORFV050 when the concentration of rifampicin increased to 250 μg/ml (Fig. 7b) and 300 μg/ml (p < 0.01), proving that rifampicin inhibits the proteolytic processing of ORFV050 at the AGA (30–32aa) putative cleavage site. At the same time, a light band of 26 kDa was observed when the concentration of rifampicin increased to 250 μg/ml, which was suspected to be the hydrolysate fraction of ORFV050 cleaved at the AG/F site.
Effect of rifampicon on ORFV050 proteolytic processing. a OFTu cells were infected with NA1/11 at MOI = 10 and treated immediately with different concentrations of rifampicin (0 l, 100, 150, 200, 250, 300, and 400 μg/ml). After 24 h of infection, western blots were performed with anti-ORFV050 polyclonal antiserum and GAPDH. b Densitometry of ORFV050 and its hydrolysate bands normalized to the level of the GAPDH control (*p < 0.05; **p < 0.01)
Discussion
ORFV replicates in the cytoplasm of host cells with various virus-encoded proteins being synthesized during maturation. During this processing, proper proteolysis is essential for virion maturation [8]. The nascent ORFV050 precursor protein is highly expressed at late stages during ORFV infection (after DNA replication). Both ORFV and VV are poxvirus, sharing analogous structures and functions in virus maturation and assembly. The ORFV050 protein has a high degree of similarity with VV P25K, playing an important role in virus maturation. Previous studies show that a lack of VV core protein VP8 may reduce the levels of steady-state RNA [17]. Assays have demonstrated that virions without VP8 have transcription rates reduced by 80–96%, compared with wild-type virus.
Previous studies showed that proteolytic processing plays important roles during replication of viruses in their host cells, mainly affecting virion maturation and viral morphogenesis. Various proteins were matured by proteolytic processing in the replication cycle of poxviruses, which is essential for correct virion assembly. As described in a study by Byrd [9], major structural proteins (4a, 4b, 25K) of VV are first synthesized as precursors and then undergo proteolytic processing to produce mature proteins (p4a, p4b, P25K) [7, 18, 19]. Most of the structural proteins required for virus assembly are expressed late in the viral infection, considering that cleavage happens at the late phases of the VV replication cycle under the action of proteinases. G1L and 17L of the VV are typical proteolytic enzymes synthesized by the infecting virus [20, 21], and are capable of selectively recognizing and cleaving particular peptide bonds of the precursor. A small AG/X motif occurs in the precursor proteins of most poxviruses, such as fowl poxvirus, VV and ORFV, and the similarity of these cleavage sites may indicate that the mechanisms for precursor proteolysis are alike [15, 22, 23].
Key features of proteolytic processing are (i) the presence of an AG/X motif for recognition by specific proteases; (ii) the proteolytic protein expressed and synthesized late during the infection; (iii) packaging into assembled virion composed of viral core particles. (iv) coexpressed with protease in both time and space [7, 24, 25]. Proteases, which play essential roles in viral core protein proteolytic processing are usually encoded by the virus itself and specifically recognize the AG/X motif. A previous study revealed that the VV P25K core protein precursor was hydrolyzed by GIL protease [20]. GIL protease is similar to ORFV037 of the ORFV in amino sequence and structure, suggesting that ORFV037 may also have a proteolytic function.
There are three putative cleavage sites in both the N-terminus and the C-terminus of the ORFV050 protein: AGF (17–19aa), AGA (30–32aa), and AGA (253–255aa). The predicted hydrolysate fractions of each motif site were 2 and 26.5, 3.5 and 25, and 28 and 0.5 kDa, respectively. In VV, virion maturation and proteolytic processing of the major structural proteins are coordinately blocked after rifampicin treatment. The proteolytic processing at AGA site of VV P25K has been proven by VanSlyke [10]. Our data, consistent with the precedent of VV P25K, indicate that AGA (30–32aa) is a putative cleavage site of ORFV050 and the 25 kDa product pORFV050 is derived from the pCDNA3.0-050 precursor. Normally, ORFV050, with a molecular mass of 28.5 kDa, is proteolytically processed into a 25 kDa fraction. According to the prediction, the AG/F site may undergo proteolytic processing with a 26 kDa fraction. However, a fraction slightly larger than 25 kDa (26 kDa) was found in OFTu cells 24 h post infection, which seemed to be the fraction of cleavage at the AG/F site (Fig. 7a). Interestingly, in the rifampicin experiment, when the concentration of rifampicin increased, the synthesis of the 25 kDa fraction was reduced, but not that of the 26 kDa fraction. It appears that cleavage at the AG/F site was not blocked by rifampicin, and, with the reduction of the 25 kDa product, the 26 kDa band can be clearly observed. We concluded that rifampicin can block only the proteolytic processing at the AG/A site, but the decrease in cleavage efficiency at the AG/S site, relative to the AG/A site, masks the presence of any intermediates. Hydrolysis at the AG/F site remains to be further investigated.
Recombinant ORFV050 protein cannot undergo proteolytic processing in the absence of ORFV because proteinase synthesis during viral replication is necessary for this processing. There are two putative ORFV proteinases, ORFV037 and ORFV035 [26], which may play key roles in the proteolytic processing of ORFV050, and need to be investigated further.
Conclusion
The current study shows that ORFV050 is a late gene product located in the cytoplasm of host cells and shares a high degree of similarity with VV P25K. With the comparison of full-length ORFV050 and site-directed mutation of pCDNA3.0-050-FLAG, we conclude that AG/A (30–32aa) is a putative cleavage site and the 25 kDa product is derived from the ORFV050 precursor. Rifampicin can inhibit the proteolytic processing of ORFV050 during the replication cycle of the virus. Additional mechanisms controlling the proteolytic processing should be investigated to develop inhibitors with potential therapeutic value in order to better control viral replication and spread of orf disease.
References
K. Zhang, Y.J. Liu, H.J. Kong, Y.J. Shang, X.T. Liu, Vector-Borne Zoonotic Dis. 14, 365–367 (2014)
H. Li, Z.Y. Ning, W.B. Hao, S.M. Zhang, X.Q. Liao, M. Li, S.H. Luo, Virus Genes 44, 429–440 (2012)
R. Cottone, M. Buttner, B. Bauer, M. Henkel, E. Hettich, H.J. Rziha, Virus Res. 56, 53–67 (1998)
C.H. Duan, M.Y. Liao, H. Wang, X.H. Luo, J. Shao, Y. Xu, W. Li, W.B. Hao, S.H. Luo, Gene 555, 260–268 (2015)
P. Lee, D.E. Hruby, J. Virol. 67, 4252–4263 (1993)
C. Lopez-Otin, C. Simon-Mateo, L. Martinez, E. Vinuela, J. BiolChem. 264, 9107–9110 (1989)
X.P. Wang, B. Xiao, J.F. Zhang, D.X. Chen, W. Li, M. Li, W.B. Hao, S.H. Luo, Front. Microbiol. 7, 538 (2016)
S.S. Whitehead, D.E. Hruby, J. Virol. 68, 7603–7608 (1994)
C.M. Byrd, D.E. Hruby, Rev. Med. Virol. 16, 187–202 (2006)
J.K. VanSlyke, C.A. Franke, D.E. Hruby, J. Gen. Virol. 72, 411–416 (1991)
W. Li, W.B. Hao, Y.Z. Peng, C.H. Duan, C.B. Tong, D.G. Song, F. Gao, M. Li, D.L. Rock, S.H. Luo, Arch. Virol. 160, 253–266 (2015)
C. Billinis, V.S. Mavrogianni, V. Spyrou, G.C. Fthenakis, Virol. J. 9, 24 (2012)
D.G. Diel, S.H. Luo, G. Delhon, Y. Peng, E.F. Flores, D.L. Rock, J. Virol. 85, 2037–2049 (2001)
D.G. Diel, G. Delhon, S.H. Luo, E.F. Flores, D.L. Rock, J. Virol. 84, 3962–3973 (2010)
S.J. Yang, D.E. Hruby, Virol. J. 4, 73 (2007)
B. Sodeik, G. Griffiths, M. Ericsson, B. Moss, R.W. Doms, J. Virol. 68, 1103–1114 (1994)
D. Wilcock, G.L. Smith, J. Virol. 70, 934–943 (1996)
J. Rosel, B. Moss, J. Virol. 56, 830–838 (1985)
R. Heljasvaara, D. Rodriguez, C. Risco, J.L. Carrascosa, M. Esteban, J.R. Rodriguez, J. Virol. 75, 5778–5795 (2001)
C.M. Byrd, T.C. Bloken, D.E. Hruby, J. Virol. 76, 8973–8976 (2002)
A.E. Aleshin, M. Drag, N. Gombosuren, G. Wei, J. Mikolajczyk, A.C. Satterthwait, A.Y. Strongin, R.C. Liddington, G.S. Salvesen, J. Biol. Chem. 287, 39470–39479 (2012)
S.J. Yang, Virol. J. 4, 78 (2007)
P. Lee, D.E. Hruby, Virology 207, 229–233 (1955)
M.D. Kazanov, Y. Igarashi, A.M. Eroshikin, P. Cieplak, B. Ratnikov, Y. Zhang, Z.W. Li, A. Godzik, A.L. Osterman, J.W. Smith, J. Proteome. Res. 10, 3642–3651 (2001)
C. Simon-Mateo, G. Andres, F. Almazan, E. Vinuela, J. Virol. 71, 5799–5804 (1997)
C.M. Byrd, D.E. Hruby, Virol. J. 2, 63 (2005)
Acknowledgements
This study was supported by the National Natural Science Foundation of China (NSFC, No. 31170147) and Guangdong Natural Science Foundation (Grant Numbers: S2013010014007 and 2014A030313070) and the Guangdong Province Science and Technology Project Plan and Social Development Foundation (2010A030400006 to Chaohui Duan).
Authors’ contribution
HW, CD, and XW participated in the design of the study. HW, CD, JJ, RD, XW, ML, JS, XL, and SL performed the experiments. HW, JJ, CD, and SL analyzed the data and wrote the manuscript. All the authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that no competing financial interests exist and no conflicts of interest exist.
Ethical standards
All animal procedures were reviewed and approved by the Institutional Animal Care and Use Committee at South China Agricultural University (The Certification Number: CNAS BL0011).
Additional information
Edited by Joachim Jakob Bugert.
Rights and permissions
About this article
Cite this article
Wang, H., Jiang, J., Ding, R. et al. Identification and characterization of Orf virus 050 protein proteolysis. Virus Genes 53, 400–409 (2017). https://doi.org/10.1007/s11262-017-1430-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-017-1430-6