Abstract
Genome-wide association studies (GWAS) allow identifying genomic regions related to traits of economic importance in animals of zootechnical interest. The objective of this research was to conduct a genome-wide association study on meat quality traits using the Illumina OvineSNPs50 BeadChip array. The animals were sampled in the departments of Córdoba, Cesar, and Valle del Cauca. The genotypes obtained with the Illumina OvineSNP50 BeadChip microarray were analyzed SNP (single-nucleotide polymorphism) data to conduct a GWAS for pH and water-holding capacity (WHC) traits measured after 7 days of maturation, in the Longissimus dorsi (LD) muscle, in 167 Creole hair sheep of 12 months old belonging to Pelibuey (CHSP, n = 60), Ethiopian (CHSE, n = 44), and Sudan (CHSS, n = 63) breeds. The GWAS was done using a mixed linear model (MLMA) and based on the Ovis aries v3.1 genome. The CHSE showed the lowest meat juice release and, consequently, the highest water-holding capacity (WHC = 30.6 ± 0.1), suggesting that this breed has better performance in the meat industry compared with CHSS (WHC = 41.7 ± 0.1) and CHSP (WHC = 36.8 ± 0.1), since there is a relationship between WHC and juiciness. For the character pH, it was not possible to annotate genes related to meat quality, while, for the WHC, they have obtained 11 candidate genes associated (ELOVL2, ARAP2, LOC101102527, SHOC2, AIPL1, CSRNP3, IFRD, KDM8, NANS, DAPK1, IBN2, TPM2). Particularly, ELOVL2, ARAP2, IBN2, and TPM2 genes are involved in muscle contraction and fatty acid composition in sheep. In this study, we generated a baseline for GWAS related to meat quality traits in Colombian Creole hair sheep that can be used for future genomic selection plans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In Colombia, Creole hair sheep (CHS) are considered a socially important genetic resource for rural economy-based communities due to the multiple benefits that these animals provide, especially, to indigenous communities (Revelo et al. 2020). Furthermore, CHS exhibit high meat production potential given the diversity of agroecosystems and favorable conditions in the country (Revelo et al. 2020; Flórez et al. 2020).
Hair sheep are mainly raised in the tropical Caribbean region under traditional production systems that lack technology and reproductive control (Flórez et al. 2020). Additionally, a group of hair sheep called “Pelibuey” (literally, sheep with cow hair) are mainly raised in southwestern Colombia (Valle del Cauca) in association with sugarcane plantations, where they provide biological control of weeds in the rows of sugarcane grass paddocks.1 According to the sheep slaughter survey reported by the National Administrative Department of Statistics (Departamento Administrativo Nacional de Estadística) (DANE 2020), 31,978 head were slaughtered, producing 1,006,355 kg of pie and 498,679 kg of carcass meat. In the same year, the national live weight at slaughter totaled 33.1 kg ± 2.5 kg, while the weight of the carcass was 18.5 kg ± 1.2 kg with a performance of 49.5%. Sheep meat consumption is internal and regionalized. The largest volume of sheep carcass meat is commercialized in market plazas (94.5%) and has grown up to 75.0%. The consumption per capita ranges from 310 to 500 g per year (Arevalo and Correa Assmus 2013).
The increasing demand of sheep meat in the country (Mestra-Vargas et al. 2019) is generating new perspectives for livestock production. However, there are limited studies on meat quality-related traits (Navarro et al. 2015) and the parameters that define these traits (e.g., water-holding capacity and pH), causing farmers to obtain low prices that are not differentiated by the quality of the meat (Arevalo and Correa Assmus 2013). Furthermore, quality-related parameters are not implemented in genetic improvement plans since WHC and pH measurement are expensive and difficult to obtain by requiring the slaughter of the selection candidates.
Warner et al. (2010) suggest that meat quality is influenced by genetic factors such as breed and type of muscle fiber. Environmental factors such as management system, diet, age, weight at sacrifice, and antemortem and postmortem procedures also affect quality (Pearce et al. 2011; Joo et al. 2013; de Lima Júnior et al. 2016; Hervé 2013). These aspects are usually considered at each stage of the meat production chain since the consumer is not only the final recipient but also the breaking point by deciding on the final product (Hervé 2013; Leal-Gutiérrez 2013).
Currently, WHC and pH traits are used to estimate meat quality (Pearce et al. 2011; Leal-Gutiérrez 2013). According to Zhang et al. (2009), WHC defines the ability of meat to retain water during the exertion of external forces such as cutting, heating, and grounding. WHC is a highly relevant trait since it affects product performance and has important economic implications on the industry and the consumer (Leal-Gutiérrez et al. 2014; Navarro et al. 2015). On the other hand, pH is one of the main parameters used to verify meat quality since it correlates with other traits including WHC and relates to color, tenderness, juiciness, and texture (Warner et al. 2010; Pearce et al. 2011; Navarro et al. 2015).
A genome-wide association study (GWAS) is a feasible and powerful tool to discover candidate genes and loci associated with quantitative traits (Yang et al. 2011; Zhao et al. 2016; Hernández-Montiel et al. 2020). It allows not only identifying controlled phenotypes for single genes but also complex traits produced by the interaction of many genes. This methodology has been used to study diseases in sheep (White et al. 2012; Leymaster et al. 2013), characteristics associated with the carcass (Palacios 2018), variations in the number of lambs per birth in Pelibuey sheep (Hernández-Montiel et al. 2020), productivity (AI-Mamun et al. 2015), growth and meat production traits (Zhao et al. 2016), resistance to parasites (Sallé et al. 2012), among others. However, to our knowledge, there are no GWAS for meat quality traits such as WHC and pH. Therefore, the objective of this research was to use SNPs to conduct a GWAS on pH and water-holding capacity (WHC) in the Longissimus dorsi muscle of Pelibuey (CHSP), Ethiopian (CHSE), and Sudan (CHSS) Creole hair sheep.
Materials and methods
Ethics statement
The sample collection procedure was approved by ethics committee of Universidad Nacional de Colombia authorized the project by which this research was conducted (in minutes No.005/2019). All procedures followed the guidelines and regulations established in the Colombian code of bioethics (resolution 8430 of 1993) and Law 84/1989 regarding the protection of animals. Blood samples were collected by qualified veterinarians during their routine practice within the framework of official programs aimed at the identification, health monitoring, and parentage confirmation of the breeds and populations included in this study. The use of animals and private land in this study was approved by the respective owners.
Animal sampling and study area
For CHSE, we evaluated 44 individuals (25 males and 19 females) from four farms located in the Department of Cordoba and one farm in the Department of Cesar (Table 1). The sheep were raised in extensive production systems and fed with Angleton (Dichantium aristatum), Brachiaria (Brachiaria sp.), Colosuana (Bothriochloa pertusa), and Guinea (Megathyrsus maximus) pastures. For CHSS, we analyzed 63 animals (42 males and 21 females) from five farms located in Cesar and one farm in Cordoba (Table 1). These animals were also raised in extensive production systems with Colosuana (Bothriochloa pertusa), Angleton (Dichantium aristatum), and Guinea (Megathyrsus maximus) pastures. For these farms, we were not able to obtain a family structure due to low technological levels. For CHSP, we evaluated 60 males from El Hatico Natural Reserve, located in the municipality of Cerrito (Valle del Cauca) (Table 1). The animals were raised in a silvopasture environment integrated with sugarcane plantations. Furthermore, the farm applies a directed breeding so we were able to obtain a family structure of 60 offspring from six fathers.
Phenotyping for pH and WHC
In total, 167 animals were slaughtered at 12 months old at different slaughterhouses according to their origin: the animals from Cordoba were slaughtered at Frigocer Expocol S.A.S. (Cereté); those from Cesar at Echeverry Gutiérrez & CIA SC (San Juan del Cesar, La Guajira), and those from Valle del Cauca at Carnes y Derivados de Occidente S.A. (Yumbo). After slaughter and at 24-h postmortem, we extracted the Longissimus dorsi (LD) muscle from the left carcass, which was vacuum sealed and stored at 4 °C. After 7 days of maturation (aging), we measured the pH using a pH meter, with three readings per sample. Additionally, the WHC was determined according to the protocol described by Leal-Gutiérrez et al. (2014). Briefly, 20 g of the LD samples was ground for 30 s, then, approximately 0.3 g of the sample was placed on a Walkman No. 5 filter paper (Figs. s1a) and a compression force of 2.5 kg was applied for 5 min. Then, the paper was removed, revealing two areas, one formed by the pressed meat (M) and the second corresponding to the water released by the meat (T) (Fig. 1. Fig.s1b). The papers were photographed using Adobe Acrobat Professional and the two areas were measured using the measure function. The WHC is expressed as the ratio between the areas M and T, and is given by the formula: Area-M*100/Area-T.
(a) Paper labeled with the number of the animal and the corresponding meat slice, (b) ground meat sample (0.3 g) on the paper after applying 2.5 kg, and (c) paper after the applied weight, showing two different areas; the inner ring is formed by the pressed meat (M) and the outer ring is formed by the water released by the meat (T), (d) delimitation of the perimeter of the two areas
DNA extraction, genotyping, and quality control
A total of 10 mL of blood from each animal was collected in BD vacuum tubes (Becton Dickinson, Franklin Lakes, NJ, USA) with k3-EDTA. Genomic DNA was extracted from 167 blood samples using the QIAamp® DNA Mini Kit (QIAGEN, Hilden, Germany). DNA quantity and quality were assessed by spectrophotometry using a ColibriTM equipment (Titertek Berthold, Neulingen, Germany) and agarose gel electrophoresis using a standard protocol to evaluate sample degradation.
All animals were genotyped with the Illumina OvineSNP50 SNP chip, which allows the simultaneous characterization of up to 54,241 independent SNPs, according to the Infinium® Assay Super II Illumina® protocol for use on the HiScan®SQ System and BeadChips scanning. Quality control (QC) was performed using PLINK v1.9b5.2 (Purcell et al. 2007) to remove SNPs with a minor allele frequency (MAF) less than 0.1 (7038), a call rate less than 0.90 (3806), and a deviation from the Hardy–Weinberg equilibrium determined at p < 0.0001 (696). Furthermore, SNPs on sex chromosomes and the mitochondrial contig were also excluded. Finally, 42,701 SNPs remained for further analyses. The genetic structure of the population of CHS was evaluated by a multidimensional scaling analysis (MDS) based on pairwise identical-by-state (IBS) distances that were calculated and plotted using Bite (Milanesi et al. 2017) package from Bioconductor in R v3.6.0 (http://www.r-project.org).
Statistical analyses
To evaluate the phenotypes for WHC and pH values, a descriptive analysis and an ANOVA of least squares or GLM were performed using the statistical package SAS 9.1.3 (SAS, Inc. Cary N.C). For the CHSE and CHSS breeds, the effects of different farms were considered, while for CHSP, controlled breeding was conducted on a single farm where 10 male offspring were produced per sire. For that reason, it was used two fixed models. First, model (M1) was used for CHSE and CHSS, where the following fixed effects were included: sex (male and female), farm (1B, 1C, 1G, 1I, 1U, 2A, 2C, 2P, 2SJ, 2VC, and 2VL), and lambing season (drought and rainy).
(M1)
where Yijkl is the vector of traits WHC and pH, μ corresponds to the overall mean, Si is the fixed effects of the i-nth sex, Fj is the fixed effects of the j-nth farm, EPk is the fixed effects of the k-nth lambing season, (SF)ij is the fixed effects of the interaction between sex and farm, (FP)jk is the fixed effects of the interaction between sex and lambing season, (SFP)ijk is the fixed effects of the interaction between farm and lambing season, and Ɛijkl is the error.
Second, model (M2) included only two individuals for CHSP and the fixed effects were as follows: father sire (1, 2, 3, 4, 5, and 6) and lambing season (drought 1 and rainy 2).
(M2)
where Yijkl is the vector of traits WHC and pH, μ corresponds to the overall mean, Pi is the fixed effects of the i-nth father, EPk is the fixed effects of the k-nth lambing season, (SF)ij is the fixed effects of the interaction between father and lambing season, and Ɛikl is the error.
Duncan’s pairwise comparisons test was used to determine significant differences, using S.A.S. 9.1.3.
GWAS analysis
Before conducting the GWAS, linear models were fitted to each phenotype to identify the fixed effects models that provided the best fit (Supplementary material). A mixed linear model (MLMA) was used to control for population structure and environmental effects. Specifically, the genetics (through the sire) of the sheep and environmental effects such as, sex, farm (M1), and lambing season (M2) were controlled. Also, the model included a genomic relationship matrix (GRM) and the variance explained by the SNPs.
The model was:
where y is the vector of phenotypes (WHC, pH), μ is the overall mean for the trait, β is the vector of fixed effects, u is the vector of random effects, X and Z are the design matrices mapping vectors of fixed or random effects, G is the genomic relationship matrix, and Ɛ is the vector of residual effects.
To facilitate this calculation, the genetic variance, var (G), is estimated based on the null model, y = μ + u + e, and is fixed while the association between each SNP and the trait is tested. In the null model, the variables indicate the following: u = ~ N (0, Gσ2), where G is the genomic relationships matrix (GRM) and σ2u = polygenic variance estimated by the null model. Table S3 shows the genomic association models (GWAS) using a MLMA for WHC and pH in CHSE, CHSS, and CHSP.
QQ-plots (Quantile–Quantile plots) were utilized to visualize the test statistics under the hypothesis of no association (null hypothesis) to identify if there was inflation.
The Bonferroni correction, which is a statistical technique to determine the threshold, was used. This technique divides the 0.05 significance level by the number of markers used in the GWAS (42,701 SNPs). Therefore, the significance threshold after applying the Bonferroni correction (1.17 × 10^ − 6) is − log10(p) ≥ 5.93, for genome-wide significance threshold. The chromosome-wide significance threshold had a value of 9.97 × 10^ − 4, which in the Manhattan plots corresponds to a value of 3.01. The Manhattan plots were generated using GWAS-tools package (Gogarten et al. 2012) in R v3.6.0 (http://www.r-project.org) to visualize the level of significance of the GWAS, the chromosomal locations, and SNPs.
Functional analysis
The functional information and biological processes of the mapped genes were retrieved using AnnotationHub, Mygene, and ReactomePA libraries (Yu 2012) of Bioconductor in R v.3.6.0, (http://www.r-project.org) by assigning functions and GO terms reported in Gene Ontology (GO), Kyoto Encyclopedia of Genes (KEGG) https://www.genome.jp/kegg/), and PATHWAY. Additionally, gene networks and gene enrichment analyses were done using the Database for Annotation, Visualization and Integrated Discovery (DAVID) and its integrated databases (INTERPRO, UP-Keywords, Uniprot, SMART, and Pir-Superfamily).
Results
Meat quality traits — WHC and pH — in three populations of Creole hair sheep
In sheep, a normal pH ranges from 5.5 to 5.8 and is related to desirable characteristics in meat quality, such as color, shear force, and water holding capacity (Arce-Recinos et al. 2021). The average pH value of the LD in CHSE was 5.75 ± 0.1 (Table 2). We did not find significant differences between sexes or farms (Table S1a) but rather for the lambing season (p < 0.01). The average WHC value of the LD in CHSE was 30.63% ± 10.9 and no significant differences were observed between sexes (Table 2, Table S1b). However, we found significant differences between farms (p < 0.05), specifically, for farm 2C. This farm is the only one located in the Department of Cesar (Table 1) in a Tropical Very Dry Forest according to Holdridge’s life zones (Palacios 2018). Furthermore, we could not evaluate 10 animals in farm 2C since it applies a low-technology production system that limited the possibility of conducting breeding control.
The average pH value of the LD in CHSS was 5.7 ± 0.1 (Table 3). Furthermore, we observed significant differences between farms (p < 0.01) and for the interaction farm*lambing season (p < 0.05, Table S1c). Moreover, for WHC of the LD in CHSS, the average value was 41.69% ± 16.3 and we determined significant differences between farms (p < 0.01) and the interaction farm*lambing season (p < 0.01, Table S1d). Farm 1U showed the lowest WHC (Table 3) and significantly differed from the other farms. This farm is located in the Department of Cordoba (Table 1) in a Tropical Dry Forest according to Holdridge’s life zones (Palacios 2018).
Finally, the average pH value of the LD in CHSP was 5.74 ± 0.1 (Table 4). We determined significant differences between fathers (p < 0.001, Table S1e); accordingly, the lowest values were observed for individuals from fathers 1 and 2. Moreover, for WHC, ram lambs showed an average value of 36.8% ± 8.8 (Table 4). We found significant differences between fathers (p < 0.001, Table S1f), and the highest values were obtained for fathers 3 (38.47% ± 9.13), 4 (41.64% ± 7), 5 (41.17% ± 7.25), and 6 (42.54% ± 7.4).
Population structure
The first two components of the bidimensional PCA plot explained 11.8% and 6.1% of the total variance, respectively. Furthermore, we observed a separation between CHSP (blue) vs CHSE (red) and CHSS (yellow) along the first axis, and a secondary split of CHSS vs CHSE along the second axis (Fig. 2). However, a slight overlap was observed between individuals of CHSS and CHSE, caused by a group of Sudan Blanco sheep (n = 8) that was sampled at La Playa farm in Valledupar. This group of sheep shares a common genetic background with CHSE (n = 4), which were sampled at Caracas farm in Valledupar. Consequently, the overlap indicates that Sudan Blanco sheep are a mix between CHSE and CHSS. We observed population stratification due to the presence of the three breeds from the different farms with contrasting genetic backgrounds. Individual kinship and population stratification in GWAS are the main causes of false positive correlations; therefore, we performed a GWAS per breed to identify candidate SNPs and genes.
Genome-wide association study (GWAS) of meat quality traits — WHC and pH
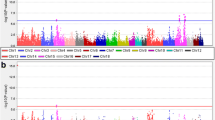
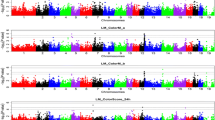
A GWAS for each of the three breeds was tested using mixed linear models (MLMA) with fixed effects. The criterion applied to select the genomic association model was based on previous results of an ANOVA on phenotypic data (Table S1a, b, c, d, e, f). The GWAS of pH did not show significantly associated SNPs. On the other hand, the GWAS of WHC for CHSE showed 12 significant SNPs located on eight chromosomes (3, 4, 6, 14, 16, 19, 20, and 21) and eight genes (Table 5). Furthermore, 25% of these SNPs were found in intergenic regions, while 66.6% were in introns and 8.3% in exons (Table 5). The Manhattan plot of WHC for CHSE is shown in Fig. 3A. For CHSS, we found 33 SNPs significantly associated with WHC (p < 0.001, Table 6), located on 16 chromosomes (1–7, 9, 11, 13, 15, 17–20, 22, and 24) and 28 genes (Table 6). The SNPs were distributed throughout intergenic regions (33.3%), introns (60.6%), promotors (3%), and exons (3%). The Manhattan plot of WHC for CHSS is shown in Fig. 3B. Finally, for CHSP, we identified nine SNPs significantly associated with WHC (p < 0.001, Table 7), located on five chromosomes (2, 5, 11, 13, and 20) and eight genes (Table 7). Furthermore, 44.44% of the SNPs were found in intergenic regions, 33.33% in introns, and 11.11% in promoters. The Manhattan plot of WHC for CHSP is shown in Fig. 3C.
Manhattan plots of WHC for three Colombian Creole-hair sheep breeds: (A) CHSE; (B); CHSS; (C) CHSP. The grey horizontal lines in the Manhattan plots indicate the significance thresholds (p < 0.001). The x-axis shows the physical positions of the SNPs per chromosome based on the Ovis aries v3.1. genome assembly
Gene ontology (GO) and KEGG gene enrichment analysis for WHC in Colombian Creole hair sheep breeds
For CHSE, 20 GO terms were assigned to the ARAP2 gene (Table S2a). Additionally, we identified three terms for the ELOVL2 gene based on KEGG functional assignments (Table S2a). Regarding CHSS, 110 GO terms were assigned to five genes (Table S2b). Most terms were assigned to the PRKCQ gene (Table S2b), followed by AIPL1, SHOC2, and LOC101102527. The functional KEGG annotation revealed seven terms related to PRKCQ and one term to LOC101102527 (Table S2b). Additionally, a pathway enrichment analysis using DAVID significantly associated the term “nucleus” (p = 5.7E − 2) to four genes, namely AIPL1, CSRNP3, IFRD1, and KDM8. Finally, for CHSP, 50 GO terms were associated with two genes (Table S2c), mostly FBN2, followed by TPM2. The KEGG annotation showed six terms associated with DAPK1, TPM2, and NANS.
Discussion
The water-holding capacity is possibly one of the most important quality traits of raw meat products (Huff-Lonergan and Lonergan 2005). Therefore, new methodologies should be implemented to improve economically important traits in the meat industry (Leal-Gutiérrez et al. 2014). We also evaluated the pH of the LD muscle since it is directly related to WHC (Pearce et al. 2011; Torrescano et al. 2009). Most water in muscle is retained in the myofibrils (Huff-Lonergan and Lonergan 2005); therefore, an accelerated decrease in pH and consequent reduction in WHC generate the Pale, Soft, and Exudative (PSE) condition of meat (Warner et al. 2010). On the other hand, high pH values result from a slow death since chronic stress for a long period before slaughter reduces glycogen reserves in the muscle and causes a low lactic acid content. For this reason, a higher pH at 24-h postmortem (6.0 to 6.8) compared to normal meat (5.4 to 5.9) turns the meat dark, firm, and dry (DFD) with a high WHC (Pearce et al. 2011), These characteristics cause dark cuts and the meat will likely be rejected by the consumer, given the impression that is not appetible or comes from old animals (Leal-Gutiérrez et al. 2014).
In this study, the pH values (CHSE = 5.75, CHSS = 5.7, CHSP = 5.74) indicated that the meats were red, firm, and non-exudative (RFN), which are optimal meat quality parameters (Gallo and Tadich 2008). This good meat quality was attributed to relevant environmental factors, such as animal manipulation and carcass management for 24 h before and after slaughter according to the established protocols. These findings demonstrate that an adequate management before and after slaughter is essential to obtain good quality meat.
The WHC is expressed as a percentage and is related to the amount of juice released by compression or manipulation; therefore, greater juice release indicates lower liquid retention and, consequently, lower WHC (Leal-Gutiérrez et al. 2014; Navarro et al. 2015). The amount and location of water in meat can vary depending on several factors related to the muscle tissue Pearce et al. 2011). Generally, in sheep, the water-holding capacity decreases with age so adult sheep meat releases a higher percentage of juice than lamb meat. However, scientific reports on the WHC of sheep meat are varied. For example, Pérez et al. (2006) indicated low values for lactating lamb of Suffolk Down × German Precocious Merino slaughtered at live weights of 10 and 15 kg, reporting a WHC of 14.5% in females and 12.5% in males. On the other hand, a WHC of 42.4 ± 2.3 (p ≤ 0.05) was reported for individuals of Pelibuey × Katadin × BlackBell that were slaughtered at an average weight of 40 kg at 10 months old (Ramírez-Bribiesca et al. 2007). We report a WHC at 7-day postmortem of 41.69% for CHSS, 30.63% for CHSE, and 36.8% for CHSP slaughtered at 12 months old. Furthermore, we found that CHSE showed lower juice release and, consequently, higher WHC, suggesting that sheep of this breed can exhibit better performance in the meat industry compared with CHSS and CHSP.
We did not find significant differences in the WHC of CHSE and CHSS between sexes, probably because these animals were managed under extensive grazing. These results agree with Expósito et al. (2003), who did not report differences in the WHC of the Longissimus dorsi muscle between sexes in Segureña sheep and attributed these findings to similarities in intramuscular fat.
Moreover, we determined significant differences in WHC for CHSE and CHSS between farms, which can be attributed to the geographic location of the farms (Table 2, Table 3). The farms in the Department of Cordoba are located in a Tropical Dry Forest, while the farms in Cesar are found in a Tropical Very Dry Forest.
For CHSP, we determined significant differences between fathers (Table 4) and these findings could be associated with the genetic traits of the father. Conversely, no differences were observed between lambing season (drought and rainy), which is possibly due to silvopasture at El Hatico natural reserve that guarantees the supply of high-quality forage year-round. In this farm, the animals graze on the sugarcane paddocks and are supplemented with balanced diets. Similarly, Ramírez-Bribiesca et al. (2007) reported a WHC of 42.4 ± 2.3 (p ≤ 0.05) in Pelibuey × Katadin × BlackBell sheep raised in an intensive system and slaughtered at an average weight of 40 kg at 10 months old. Moreover, Frías et al. (2011) reported a WHC of 12.8 ± 0.16 in Pelibuey with Katahdin and Dorper lambs fed with grass and supplemented with fermented sugarcane, which were sacrificed at 20 kg of weight.
Regarding pH, we did not find significant differences between sexes for CHSE and CHSS. These results agree with Rodríguez et al. (2011), who reported a pH of 5.65 and 5.61 at 24-h postmortem in the Longissimus lumborum and semimembranosus muscles in Assaf and Merino × Assaf sheep. Moreover, the effect of farm on pH was significant for CHSS and the highest values were observed in farms 2VC (5.81 ± 0.05) and 2P (5.85 ± 0.09). Conversely, we did not find significant effects of farm on pH for CHSE. For this breed, the pH values ranged from 5.7 to 5.8 and corresponded to the optimal values for meat quality. These findings are attributed to an adequate management of the animals before and after slaughter, according to standardized protocols.
SNP markers associated with WHC, gene ontology, and metabolic pathways
For CHSE, we found two SNPs potentially associated with WHC. The first SNP, OAR20_48109414.1, is located in the ELOVL2 (fatty acid elongation) gene, which encodes the enzyme that catalyzes the synthesis of long-chain polyunsaturated fatty acids (C20- and C22-PUFA), acting specifically on polyunsaturated acyl-CoA and with greater activity towards acyl-CoA C20: 4 (n-6). This enzyme participates in the production of polyunsaturated VLCFA of different chain lengths that are involved in several biological processes as membrane lipid precursors and lipid mediators (UniProt Consortium 2015). Similarly, Bolormaa et al. (2016) found SNPs strongly associated with polyunsaturated fatty acid concentrations, such as OAR20_44.1, located at 100 Kb from the ELOVL2 gene, in a GWAS of 56 traits (e.g., weight, fat, muscling, tenderness, meat color, pH level, and fatty acid profile) in 10,613 in nine sheep breeds (Merino, Poll Dorset, Border Leicester, Suffolk, White Suffolk, Texel, Corriedale, Coopworth, and various crosses). ELOVL2, along with PNPLA3 and FADS2 genes, participate in fatty acid synthesis and metabolism. Moreover, the EVOVL6 gene was found as a candidate affecting the composition of palmitic and palmitoleic fatty acids (Bolormaa et al. 2016). These two genes were also found in the dorsal fat of pigs Corominas et al. 2013).
The second candidate SNP found in CHSE is OAR6_ s36343.1, which is located in the ARAP2 gene and encodes the Arf-GAP with Rho-GAP domain, ANK repeat, and PH domain-containing protein 2. This protein modulates actin cytoskeleton remodeling by regulating ARF and RHO family members and is activated by binding to phosphatidylinositol 3,4,5-trisphosphate (PtdIns 3,4,5 P3) (UniProt Consortium 2015). Bolormaa et al. (2016) also identified ARAP2 as a candidate gene associated with meat color and concentration of polyunsaturated fatty acids (omega-3 and omega-6).
For CHSS, we propose eight candidate SNPs, including five that are associated with WHC, namely s55275.1 located in PRKCQ; OAR11_26036763.1 in AIPL1; OAR2_151938109.1 in CSRNP3; OAR4_59276586.1 in IFRD1; and OAR24_27265846.1 in KDM8. Altogether, these genes participate in the metabolic pathway of “nucleus processes.” Particularly, PRKCQ encodes a protein kinase C theta type, which belongs to the family of serine/threonine-specific kinase proteins that are activated by calcium and diacylglycerol. These protein family members phosphorylate a wide variety of protein targets and are involved in several cell signaling pathways (UniProt Consortium 2015). We found that this gene is associated with several terms involving immune-related functions, as well as with the term adipocytokine signaling pathway (oas04920) (see Supplementary Table S2c). The IFRD1 gene encodes the interferon-related developmental regulator 1 and can function as a transcriptional coactivator that controls the growth and differentiation of specific cell types during embryonic development and tissue regeneration (Stelzer et al. 2016). In sheep, this gene is associated with myoblast growth and muscle differentiation (Cheng et al. 2020). Moreover, KDM8 encodes a bifunctional enzyme that acts as an endopeptidase and a 2-oxoglutarate-dependent monooxygenase (Liu et al. 2016). The photoreceptor/pineal-expressed gene, AIPL1, encodes the arylhydrocarbon interacting protein-like 1 and is found in the candidate LCA4 region. This encoded protein contains three tetratricopeptide motifs, compatible with nuclear transport or chaperone activity (Stelzer et al. 2016). Finally, SNP OAR24_21411103.1 located in LOC101102527, SNP OAR22_34955585_X.1 in SHOC2, and SNP OAR2_151938109.1 in CSRNP3 are not reported in the literature so their functions related to WHC are unknown.
For CHSP, we found nine SNPs, including four associated with WHS. First, SNP s11926.1 is located in the DAPK1 gene that encodes a calcium-dependent threonine kinase protein. This protein is involved in several cellular signaling pathways that activate cell survival, apoptosis, and autophagy; specifically, regulating type I apoptotic and type II autophagic cell death signals, depending on the cellular setting (UniProt Consortium 2015). In this study, this gene was found associated with the term autophagy — animal (oas04140) (Table S2c). Singh et al. (2016) found it related to programmed cell death or apoptosis. Second, SNP s16895.1 is found in FBN2, which encodes the fibrilin-2 protein involved in the formation of a binding network with other proteins to produce filiform filaments called microfibrils in the muscle. The microfibrils are a component of elastic fibers that allow the skin, ligaments, and blood vessels to stretch (Frédéric et al. 2009). Researchers have suggested that fibrilin-2 plays a role in the direction of assembly of elastic fibers during embryonic development. Microfibrils also contribute to the establishment of more rigid tissues that support the crystalline lens, nerves, and muscles; furthermore, they contain the transforming growth factor beta (TGF-β) that inactivates them. When the microfibrils are released, TGF-β is activated and affects tissue growth and repairment throughout the organism (Frédéric et al. 2009; Ramirez & Dietz 2007). On the other hand, SNP OAR2_56383269.1 is located in the TPM2 gene that regulates the production of beta (β)-tropomyosin, which regulates muscle fiber tension (muscular contraction) by controlling myosin and actin binding. In non-muscle cells, tropomyosin proteins play a role in controlling cell shape (Tajsharghi et al. 2007; Ochala et al. 2007). β-tropomyosin is mainly found in skeletal muscles, which are used for movement. This protein helps to regulate muscle contraction by interacting with other muscle proteins, particularly, myosin and actin. These interactions are essential to stabilize and maintain sarcomeres within muscle cells (Tajsharghi et al. 2007; Ochala et al. 2007). Sarcomeres are the basic units of muscle contraction and they are composed of proteins that exert the mechanical force needed for muscle contraction. Noce et al. (2018) determined that TPM2, as well as ACTA1, MYLPF, MYH2, MYH7, TPM2, and TTN, encode myofibril proteins involved in muscle contraction in the Longissimus dorsi muscle in autochthonous Spanish sheep breeds (Canaria de Pelo, Roja Mallorquina, Gallega, Xisqueta, and Ripollesa). Regarding metabolic pathways associated with striated muscle contraction, we found that gluconeogenesis, glycolysis, the citric acid cycle, and the electron transport chain were enriched. Finally, SNP OAR2_56383269.1 is located in the NANS gene that encodes an enzyme involved in sialic acid biosynthesis. This protein uses N-Acetyl-mannosamine 6-phosphate and mannose 6-phosphate as substrates to produce phosphorylated forms of N-acetylneuraminic acid (Neu5Ac) and 2-keto-3-deoxy-D-glycero-D-galacto-nononic acid (KDN) (Stelzer et al. 2016).
Conclusion
We did not find SNPs significantly associated with pH probably since the measurements were taken at 7-day postmortem, when most biochemical and physical processes are stable. Therefore, this trait must be evaluated at the moment of slaughter and within 3-h postmortem to determine the velocity at which the pH decreases.
We generated a baseline for sheep farmers who seek to improve meat and carcass quality based on a genome-wide association study of meat quality traits in Colombian Creole hair sheep. We found that the LD muscle of CHSE releases fewer juice and, consequently, has greater WHC, suggesting that these animals will exhibit better performance in the meat industry. Overall, we identified candidate genes that are promising for genomic selection (ELOVL2, ARAP2, FBN2, and TPM2) and are involved in muscle contraction, metabolism, and fatty acid composition.
Data availability
The data sets generated or analyzed during the current study, or both, are available from the corresponding author upon reasonable request.
References
AI-Mamun, HA, Kwan, P, Clark, SA, Ferdosi, MH, Tellam, R. 2015. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet. Sel. Evol. 47:66. https://doi.org/10.1186/s12711-015-0142-4.
Arce-Recinos, C., Chay-Canul, A. J., Alarcón-Zúñiga, B., Ramos-Juárez, J. A., Vargas-Villamil, L. M., Aranda-Ibáñez, E. M., ... & Lopes Dias da Costa, R. (2021). Índices de eficiencia alimenticia en ovinos de pelo: calidad de la carne y genes asociados. Revisión. Revista mexicana de ciencias pecuarias, 12(2), 523–552
Arevalo, G. Á., & Correa Assmus, G. 2013. Tecnología en la ovinocultura colombiana: estado del arte. Revista ciencia animal, 1(6), 125-142.
Bolormaa, S., Hayes, B. J., van der Werf, J. H., Pethick, D., Goddard, M. E., & Daetwyler, H. D. 2016. Detailed phenotyping identifies genes with pleiotropic effects on body composition. BMC genomics, 17(1), 1-21.
Corominas, J., Ramayo-Caldas, Y., Puig-Oliveras, A., Pérez-Montarelo, D., Noguera, J. L., Folch, J. M., & Ballester, M. (2013). Polymorphism in the ELOVL6 gene is associated with a major QTL effect on fatty acid composition in pigs. PloS one, 8(1), e53687.
Cheng, S., Wang, X., Zhang, Q., He, Y., Zhang, X., Yang, L., & Shi, J. 2020. Comparative transcriptome analysis identifying the different molecular genetic markers related to production performance and meat quality in Longissimus dorsi tissues of MG× STH and STH sheep. Genes, 11(2), 183.
DANE. Boletín técnico Encuesta de sacrificio de ganado. (2020). http://www.dane.gov.co/files/investigaciones/boletines/sacrificio/bol_sacrif_IVtrim14.pdf
de Lima Júnior, D. M., de Carvalho, F. F., da Silva, F. J., Rangel, A. H. D. N., Novaes, L. P., & Difante, G. D. S. 2016. Intrinsic factors affecting sheep meat quality: a review. Revista Colombiana de Ciencias Pecuarias, 29(1), 03-15.doi: https://doi.org/10.17533/udea.rccp.v29n1a01
Expósito, C., Blanco, T. P., Peinado, J. M., García, V. D., Aldea, M. A., Martínez, A. G., ... & De La Cruz, R. A. (2003). Calidad de la canal y de la carne en corderos ligeros de raza Segureña. Archivos de zootecnia, 52(199), 315–326.
Flórez, M. J., Hernández, P. M., Bustamante, Y. M., & Vergara, G. O. 2020. Caracterización morfológica y faneróptica de hembras Ovino de Pelo Criollo Colombiano “CHS” Sudán. MVZ Cordoba, 84–93. https://doi.org/10.21897/rmvz.125 (2020).
Frédéric, M. Y., Monino, C., Marschall, C., Hamroun, D., Faivre, L., Jondeau, G., & Collod‐Béroud, G. 2009. The FBN2 gene: new mutations, locus‐specific database (Universal Mutation Database FBN2), and genotype‐phenotype correlations. Human mutation, 30(2), 181-190.
Frías, J. C., Aranda, E. M., Ramos, J. A., Vázquez, C., & Díaz, P. 2011. Calidad y rendimiento en canal de corderos en pastoreo suplementados con caña de azúcar fermentada. Avances en Investigación Agropecuaria, 15(3), 33-44.
Gallo, C., & Tadich, N. 2008. Bienestar animal y calidad de carne durante los manejos previos al faenamiento en bovinos. REDVET. Revista electrónica de Veterinaria, 9(10B).
Gogarten, S. M., Bhangale, T., Conomos, M. P., Laurie, C. A., McHugh, C. P., Painter, I., ... & Laurie, C. C. 2012. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics, 28(24), 3329–3331.
Hernández-Montiel, W., Martínez-Núñez, M. A., Ramón-Ugalde, J. P., Román-Ponce, S. I., Calderón-Chagoya, R., & Zamora-Bustillos, R. 2020. Genome-wide association study reveals candidate genes for litter size traits in pelibuey sheep. Animals, 10(3), 434.
Hervé, M. 2013. Carne ovina: Producción, características y oportunidades en lo que hoy demanda el consumidor nacional e internacional. Oficina de Estudios y Políticas Agrarias (ODEPA), Santiago, Chile.
Huff-Lonergan, E., & Lonergan, S. M. 2005. Mechanisms of water-holding capacity of meat: The role of postmortem biochemical and structural changes. Meat science, 71(1), 194-204.
Joo, S. T., Kim, G. D., Hwang, Y. H., & Ryu, Y. C. 2013. Control of fresh meat quality through manipulation of muscle fiber characteristics. Meat science, 95(4), 828-836.
Leal-Gutiérrez, J. D. 2013. Marcadores moleculares asociados a la Capacidad de Retención de Agua (CRA) en carne de Bos indicus y sus cruces (Doctoral dissertation).
Leal-Gutiérrez, J. D., Jiménez-Robayo, L. M., Ariza, M., Manrique, C., López, J., Martínez, C., ... & Jiménez, A. 2014. Efecto del tipo genético y la maduración sobre la retención de agua en carne de toros castrados. Archivos de zootecnia, 63(243), 409–418.
Leymaster, K. A., Chitko-McKown, C. G., Clawson, M. L., Harhay, G. P., & Heaton, M. P. 2013. Effects of TMEM154 haplotypes 1 and 3 on susceptibility to ovine progressive pneumonia virus following natural exposure in sheep. Journal of Animal Science, 91(11), 5114-5121.
Liu, H., Wang, C., Lee, S., Deng, Y., Wither, M., Oh, S., ... & Zhang, G. 2016. Clipping of arginine-methylated histone tails by JMJD5 and JMJD7. Proceedings of the National Academy of Sciences, 114(37), E7717-E7726.
Mestra-Vargas, L. I., Martínez-Reina, A. M., & Santana-Rodríguez, M. O. 2019. Technical and economic characterization of lamb meat production in Córdoba, Colombia. Agronomía Mesoamericana, 30(3), 871-884. http://dx.doi.org/https://doi.org/10.15517/am.v30i3.36931.
Milanesi, M., Capomaccio, S., Vajana, E., Bomba, L., Garcia, J. F., Ajmone-Marsan, P., & Colli, L. (2017). BITE: an R package for biodiversity analyses. bioRxiv.: 181610. Preprint, 29.
Navarro, Y.T., Mendoza, M.A., & Osorio, D. S. D. 2015. Influencia de la edad y el tiempo de madurez en la capacidada de retencion de agua (CRA) en la carne de ovino criollo. @ limentech, Ciencia y Tecnología Alimentaria, 13(1), 41–47.
Noce, A., Cardoso, T. F., Manunza, A., Martínez, A., Cánovas, A., Pons, A., ... & Amills, M. 2018. Expression patterns and genetic variation of the ovine skeletal muscle transcriptome of sheep from five Spanish meat breeds. Scientific reports, 8(1), 1–7.
Ochala, J., Li, M., Tajsharghi, H., Kimber, E., Tulinius, M., Oldfors, A., & Larsson, L. 2007. Effects of a R133W β‐tropomyosin mutation on regulation of muscle contraction in single human muscle fibres. The Journal of physiology, 581(3), 1283-1292.
Palacios, Y. 2018. Análisis genómico de características de crecimiento muscular y de calidad de la canal en poblaciones ovinas de pelo. Universidad de Colombia.
Pearce, K. L., Rosenvold, K., Andersen, H. J., & Hopkins, D. L. 2011. Water distribution and mobility in meat during the conversion of muscle to meat and ageing and the impacts on fresh meat quality attributes A review. Meat science, 89(2), 111-124.
Pérez, P., Maino, M., Tomic, G., Köbrich, C., Morales, M. S., & Pokniak, J. 2006. Calidad de carne de corderos lechales del cruce Suffolk Down x Merino Precoz Alemán: Efecto del peso de sacrificio y sexo. Archivos de Zootecnia, 55(210), 171-182.
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., ... & Sham, P. C. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American journal of human genetics, 81(3), 559–575. https://doi.org/10.1086/519795.
Ramírez-Bribiesca, E., Hernández-Cruz. L., Guerrero-Legarreta, I., & Hernández-Calva, L.M. 2007. Calidad de la carne y análisis sensorial en ovinos de pelo y lana provenientes de engorda intensiva en méxico. Vº Congreso de Especialistas en Pequeños Rumiantes y Camélidos Sudamericanos, Mendoza, Argentina.
Ramirez, F., & Dietz, H. C. 2007. Fibrillin‐rich microfibrils: Structural determinants of morphogenetic and homeostatic events. Journal of cellular physiology, 213(2), 326-330.
Revelo, H. A., López-Alvarez, D., Landi, V., Rizzo, L., & Alvarez, L. A. 2020. Mitochondrial DNA Variations in Colombian Creole Sheep Confirm an Iberian Origin and Shed Light on the Dynamics of Introduction Events of African Genotypes. Animals, 10(9), 1594.
Rodríguez, A. B., Bodas, R., Landa, R., López-Campos, Ó., Mantecón, A. R., & Giráldez, F. J. 2011. Animal performance, carcass traits and meat characteristics of Assaf and Merino× Assaf growing lambs. Livestock Science, 138(1-3), 13-19.
Sallé, G., Jacquiet, P., Gruner, L., Cortet, J., Sauvé, C., Prévot, F., ... & Moreno, C. R. 2012. A genome scan for QTL affecting resistance to Haemonchus contortus in sheep. Journal of animal science, 90(13), 4690–4705.
Singh, P., Ravanan, P., & Talwar, P. (2016). Death associated protein kinase 1 (DAPK1): a regulator of apoptosis and autophagy. Frontiers in molecular neuroscience, 9, 46.
Stelzer, G., Rosen, N., Plaschkes, I., Zimmerman, S., Twik, M., Fishilevich, S., ... & Lancet, D. (2016). The GeneCards suite: from gene data mining to disease genome sequence analyses. Current protocols in bioinformatics, 54(1), 1–30
Tajsharghi, H., Ohlsson, M., Lindberg, C., & Oldfors, A. 2007. Congenital myopathy with nemaline rods and cap structures caused by a mutation in the β-tropomyosin gene (TPM2). Archives of neurology, 64(9), 1334-1338.
Torrescano, G.R., Sánchez-Escalante, A., Peñúñuri-Molina, F. J., Velázquez-Caudillo, J., & Tineo-Sierra, R. 2009. Características de la canal y calidad de la carne de ovinos pelibuey, engordados en hermosillo, sonora. Biotecnia, 11(1), 41-50.
UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Research, 43(D1), D204-D212.
Warner, R.D., Greenwood, P. L., Pethick, D. W., & Ferguson, D. M. 2010. Genetic and environmental effects on meat quality. Meat science, 86(1), 171-183. doi:https://doi.org/10.1016/j.meatsci.2010.04.042.
White, S. N., Mousel, M. R., Herrmann-Hoesing, L. M., Reynolds, J. O., Leymaster, K. A., Neibergs, H. L., ... & Knowles, D. P. 2012. Genome-wide association identifies multiple genomic regions associated with susceptibility to and control of ovine lentivirus. PLoS ONE 7(10): e47829.https://doi.org/10.1371/journal.pone.0047829
Yang, J., Lee, S. H., Goddard, M. E., & Visscher, P. M. (2011). GCTA: a tool for genome-wide complex trait analysis. The American Journal of Human Genetics, 88(1), 76-82.
Yu, G. 2012. Reactome pathway analysis. R package version 1.10.1.
Zhang, L., Yue, H. Y., Zhang, H. J., Xu, L., Wu, S. G., Yan, H. J., ... & Qi, G. H. 2009. Transport stress in broilers: I. Blood metabolism, glycolytic potential, and meat quality. Poultry science, 88(10), 2033-2041.
Zhao, F. P., Wei, C. H., Zhang, L., Liu, J. S., Wang, G. K., Tao, Z. E. N. G., & Du, L. X. (2016). A genome scan of recent positive selection signatures in three sheep populations. Journal of Integrative Agriculture, 15(1), 162-174.
Acknowledgements
The authors would like to thank the sheep farmers who contributed to this work, to Asovinos, el Hatico Natural Reserve, and small-scale producers of the departments of Cesar and Cordoba for their collaboration. We also would like to thank Andrea Gonzalez for translating the manuscript.
Funding
Open Access funding provided by Colombia Consortium. This work was supported by COLCIENCIAS, Grant Number 757 Programa de Becas Doctorales Colombia 2016.
Author information
Authors and Affiliations
Contributions
Conceptualization: LAA, MFA, ODV. Data curation: HAR and DLA. Formal analysis: HAR and DLA. Funding acquisition: LAA, HAR, and YAP. Investigation methodology: HAR, YAP, and DLA. Supervision: LAA and DLA. Visualization: HAR, LAA, and DLA. Writing—original draft: HAR and DLA. Writing—review editing: all authors.
Corresponding authors
Ethics declarations
Ethics approval
This work had the endorsement of the ethics committee of Universida Nacional de Colombia sede Palmira. Collection of blood samples was carried out by veterinarians adhering to the regulations and guidelines on animal husbandry and welfare.
Consent for publication
All the authors consent to publish the manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Revelo, H.A., López-Alvarez, D., Palacios, Y.A. et al. Genome-wide association study reveals candidate genes for traits related to meat quality in Colombian Creole hair sheep. Trop Anim Health Prod 55, 357 (2023). https://doi.org/10.1007/s11250-023-03688-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11250-023-03688-z