Abstract
Scrapie is a naturally occurring transmissible spongiform encephalopathy in sheep and goats. Resistance or susceptibility of small ruminants to classical scrapie is influenced by polymorphisms in the prion protein gene (PRNP). PRNP variability in Indonesian indigenous goat breeds has not been investigated so far and therefore was the goal of this study. Sanger sequencing of the PRNP gene coding region in 72 goats of the seven Indonesian breeds Kacang, Gembrong, Samosir, Kejobong, Benggala, Jawarandu, and Peranakan Etawah revealed three amino acid substitutions, namely W102G, H143R, and S240P. Some silent mutations were also found at codons 42 (a/g), 138 (c/t), and 179 (g/t). The PRNP alleles K222 and D/S146 known to have significant protective effects on resistance to classical scrapie in goats were not detected. The allele R143, which may have a moderate protective effect, had a frequency of 12% among the analyzed Indonesian goat breeds. While R143 was missing in Kacang and Benggala, its frequency was highest in the breed Gembrong (32%). No scrapie cases have been reported in Indonesia until now. However, in the case that selection for protective PRNP variants would become a breeding goal, the analyzed breeds will not be very useful resources. Other goat breeds which are present in the country should be investigated regarding resistance to scrapie, too.
Similar content being viewed by others
Introduction
Scrapie belongs to the transmissible spongiform encephalopathies (TSEs) like other similar diseases such as bovine spongiform encephalopathy (BSE) and human Creutzfeldt-Jakob-disease (CJD). TSEs are fatal degenerative disorders of the central nervous system, which are characterized by the accumulation of an abnormal isoform (PrPsc) of the cellular prion protein (PrPc) (Prusiner and DeArmond 1994). Scrapie has long been known in sheep and goats, with a less frequent occurrence in the latter (Chelle 1942; EFSA BIOHAZ Panel 2014). In 1998, a new, atypical type of scrapie was identified in sheep that differs in its epidemiological and biochemical characteristics from the long-known classical scrapie type (Benestad et al. 2003).
Compared to other species, the prion protein gene (PRNP) of sheep and goats is highly polymorphic, and PRNP variants, mostly amino acid substitutions, are associated with resistance or susceptibility to classical scrapie (overview given by Goldmann 2008). About 15 years ago, the European Union (EU), as well as other countries, started to select for sheep carrying the PRNP haplotype A136R154R171 (ARR), which was regarded to be resistant to classical scrapie, and to eliminate the other haplotypes, especially VRQ, which was suspected to be most susceptible (Dawson et al. 1998; Dubois et al. 2002; Baylis et al. 2004; Vitezica et al. 2005). In the EU, related breeding program requirements were integrated into several regulations. Unfortunately, the PRNP variants used in breeding do not protect sheep against atypical scrapie (Lühken et al. 2007).
In the past, scientific evidence regarding the association between PRNP variants in goats and resistance or susceptibility to scrapie has been insufficient to pursue a strategy similar to that used in sheep, particularly due to the lower number of scrapie cases in goats compared to sheep (Vaccari et al. 2009). However, in the last decade, further scientific evidence was obtained on this subject, which was evaluated by the European Food Safety Authority (EFSA). In this report, it was concluded that the results of various studies indicate that nine amino acid substitutions (codons 127, 142, 143, 145, 146, 154, 211, and 222) provide some degree of resistance to classical scrapie. Both the quality and reliability of the available field and experimental data were considered robust enough to conclude that the K222, D146, and S146 alleles confer significant genetic resistance to classical scrapie strains naturally occurring in the EU goat population. The authors of this large meta-analysis recommend to consider breeding for resistance as an effective tool for controlling classical scrapie in goats (EFSA BIOHAZ Panel 2017). A strong protective effect of S146 and K222 was already confirmed for goats heterozygous with one of these alleles, surviving oral inoculation with classical scrapie for more than 6 years, while non-carriers of these alleles showed clinical symptoms after an average of 2 years after inoculation (Cinar et al. 2018).
Unfortunately, the protective PRNP allele K222 was found only at low frequencies in the cosmopolitan goat breeds Alpine and Saanen in various countries like France, Italy, Spain, the Netherlands, the UK, and the USA. In contrast, higher frequencies of the protective allele D146 were observed in populations of the also worldwide distributed Boer goat in the Netherlands, the UK, and the USA. For the Toggenburg breed, a higher frequency of K222 was determined in the Netherlands, but not in the UK and the USA, which is maybe due to founder effects (Acutis et al. 2008; White et al. 2008; Barillet et al. 2009; Goldmann et al. 2011, 2016; Acín et al. 2013; Windig et al. 2016).
Higher frequencies of K222 or S/D146 were found in single local breeds, as, e.g., in the Garganica breed of southern Italy (Acutis et al. 2008), in the Small East African breed from Tanzania (Kipanyula et al. 2014), and in Damascus related breeds from Cyprus and Turkey (Meydan et al. 2017; EFSA 2012). These findings aroused interest in studying the PRNP variability of other local goat breeds from other regions of the world.
For Asian and Eurasian goat populations, e.g., from China, Japan, Korea, Turkey, and Pakistan, data on frequencies of PRNP polymorphisms are available (Kurosaki et al. 2005; Babar et al. 2008; Hussain et al. 2011; Zhou et al. 2013; Hassan et al. 2016; Kim et al. 2019; Meydan et al. 2017). In contrast, even since Indonesia represents a large southern part of the Asian continent, no data on PRNP variability of the Indonesian goat population has been published until now. Therefore, the aim of this study was to analyze for the first time the PRNP coding region of seven Indonesian goat breeds in order to assess their susceptibility status toward classical scrapie as well as their potential to be a resource for protective PRNP alleles.
Materials and methods
Collection of animal samples
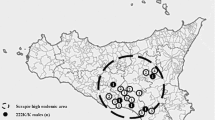
A total of 72 samples (10–11 samples per breed) from goats belonging to seven breeds were collected from different farms in several provinces in Indonesia (Fig. 1). The samples were collected in the regions in which these breeds usually are bred.
Locations of sampling Indonesian goat breeds. The map was created using ArcMap version 10.5. The OpenStreetMap provides free geographic data and is available under the Open Database Licence (https://www.openstreetmap.org/copyright)
These Indonesian goat breeds included the three native breeds Kacang (Central Java), Gembrong (Bali Island), and Samosir (North Sumatera), as well as the cross breeds Kejobong (Central Java), Benggala (Nusa Tenggara Timur), Jawarandu, and Peranakan Etawah (both Yogyakarta). In the individual farms with herd sizes of about 20–30 animals, 3–5 unrelated animals were sampled each. Two-thirds of the sampled goats were female. Depending on the management system, animals were kept in groups on pasture or indoors separated from each other. Only apparently healthy animals with a good body condition were sampled. Blood samples were collected from the jugular vein using 3 ml EDTA monovettes. All individuals were identified based on morphological characteristics and were considered to be genetically unrelated.
DNA extraction, PCR amplification, and sequencing of PRNP
Genomic DNA was isolated using a commercial extraction kit (Blood DNA isolation kit, Geneaid Biotech Ltd. New Taipei City, Taiwan) as recommended by the manufacturer. Quantity and quality of extracted DNA were checked using a NanoDrop 2000 spectrophotometer (VWR, Darmstadt, Germany). Polymerase chain reaction (PCR) for the amplification of a 970 bp fragment containing the complete PRNP coding region was done as described in Lühken et al. (2007) for ovine PRNP. Due to differences between sheep and goat PRNP sequences in the binding region of the forward primer, it was necessary to modify the forward primer (TGCCASTGCTATGCAGTCATT, changed nucleotides underlined). PCR program: An initial denaturing stage at 95 °C for 5 min was followed by 35 cycles at 94 °C for 30 s, 55 °C for 90 s, and 72 °C for 60 s, with a final step at 72 °C for 10 min. The PCR products were purified using a commercial kit (MSB Spin PCRapace, Stratec Molecular, Berlin, Germany), diluted to 10 ng/µl after quantification by agarose gel electrophoresis and sent to LGC Genomics GmbH (Berlin, Germany) for Sanger sequencing using the reverse PCR primer. The resulting chromatograms were analyzed with Chromas 1.74 (Technelysium Pty Ltd, South Brisbane, Australia). The GenBank sequence EF192309.2 was used as a reference sequence of the goat PRNP gene, representing the wild type (Acutis et al. 2008).
Calculation of genotype and allele frequencies: chi-square/Fisher’s exact tests
The genotype and allele frequencies of the identified PRNP variants were calculated based on the numbers of animals with a certain genotype of a variable PRNP position divided by the total number of animals, or the numbers of chromosomes with a certain allele of a variable PRNP position divided by the total number of chromosomes, respectively. Calculations were done over all samples as well as within single breeds. Chi-square or, in the case of numbers below five, Fisher’s exact test was used to test for significant (p ≤ 0.05) differences in the distribution of alleles between pairs of breeds.
Results
Six polymorphisms were observed in the coding PRNP region of the analyzed indigenous Indonesian goat breeds, namely the three amino acid substitutions W102G, H143R, and P240S, as well as the three synonymous mutations P42P, S138S, and V179V. All seven breeds were polymorphic for codons 42, 138, and 240, whereas codons 143, 179, and 102 were only variable in five (GEM, JAW, KEJ, PE, SAM), three (JAW, KEJ, PE), and a single breed (BEG), respectively. The W102G polymorphism occurred only in the BEG breed. Detailed genotype and allele frequencies of all identified variants over all breeds and within single breeds are shown in Tables 1 and 2. For all polymorphic positions, alleles were significantly different distributed at least between a single pair of breeds (P240S) up to seven pairs of breeds (H143R and V197V). All results of chi-square or Fisher’s exact tests are shown in Table S2.
Discussion
As in many other countries all over the world, the occurrence of scrapie is not monitored in Indonesia. The Indonesian goat population includes many native, crossbred, and foreign breeds (Pakpahan et al. 2016). To our knowledge, this is the first study on variability of the coding region of the PRNP gene in Indonesian goat breeds. By sequencing the target region in 72 goats of seven different breeds, in total six polymorphic positions were detected, three of them causing amino acid substitutions. In all analyzed breeds, at least three PRNP codons were variable, always including the amino acid substitution at codon 240.
Among the PRNP variants detected in Indonesian goats, only R143 belongs to the nine PRNP variants which are supposed to confer some degree of resistance to classical scrapie. In the analyzed Indonesian goat breeds, the R143 allele was identified at a frequency of 12% among all breeds, ranging from 0% in two breeds up to 32% in GEM. Epidemiological data from Italy, France, Greece, and Cyprus indicated an association of the 143R allele with lower susceptibility to classical scrapie (Billinis et al. 2002; Acutis et al. 2006; Vaccari et al. 2006; Papasavva-Stylianou et al. 2011; Fragkiadaki et al. 2011). For example, Billinis et al. (2002) observed a significantly higher proportion of scrapie-positive goats among those carrying the genotype HH at position 143 compared to goats with genotypes HR or RR (P = 0.012). However, 143R did not show a protective effect in vitro conversion experiments (Eiden et al. 2011). Until now, results from experimental inoculation with classical scrapie have only been reported for a single 143HR goat (Goldmann et al. 1996). According to this, R143 confers a slightly longer incubation period compared to H143, but a significantly shorter incubation period than goats with M142 (together with H143). Considering all available data, goats with R143 may be better protected compared to wild-type animals, but the potential protective effect against classical scrapie seems to be much lower compared to other PRNP alleles, especially K222 and D/S146 (EFSA BIOHAZ Panel 2017).
As a further amino acid substitution in PRNP, W102G was identified in only one of the Indonesian breeds (BEG). To our knowledge, G102 is a very rare allele, and therefore there is not enough epidemiological field data available to study a potential protective effect of G102 against scrapie in goats. An experimentally scrapie-infected goat heterozygous for three octapeptide repeats in combination with G102 (while the other chromosome carried the wild type, which is five octapeptide repeats, in combination with W102) succumbed to scrapie only after a significantly prolonged incubation period compared with goats homozygous for the wild-type allele combination (Goldmann et al. 1998). However, it is not clear if this could be attributed to the missing of two octapeptide repeat sequences, to the amino acid substitution at position 102, or a combined effect of both mutations. The loss of two octapeptides, as well as the amino acid substitution at codon 102 probably, modulates the copper ion binding function of the prion protein (Stöckel et al. 1998). Therefore, a theoretical effect on scrapie susceptibility of both of the two types of mutations could be related to the same biochemical mechanism. Finally, P240S was observed in all analyzed breeds as the third and last amino acid substitution in the prion protein of Indonesian goats. In all investigated goat populations so far, P and S at position 240 seem to be more or less equally distributed. This is also reflected in the Indonesian goat breeds, with a slightly higher frequency of P in some breeds and of S in others. No association between the length of disease incubation and the different codon 240 genotypes was found in scrapie inoculation experiments (Goldmann et al. 1996). Moreover, it is likely that, as has been demonstrated for rodent PrP (Stahl et al. 1990), the C-terminal region of goat PrP, including amino acid 240, is removed during the post-translational attachment of a glycoinositol phospholipid tail.
Taken together, in the analyzed Indonesian goat breeds, three amino acid substitutions have been identified (W102G, H143R, S240P). Only the R143 allele may have a moderate effect on resistance against classical scrapie in goats (EFSA BIOHAZ Panel 2017). The R143 allele is also present in some goat breeds in other Asian/Eurasian countries, e.g., at a frequency of about 15% in the Pakistani breeds Kamori and Local Hairy, and 29% and 37% in southern and northern Chinese goat populations, respectively (Zhou et al. 2013). The PRNP alleles with known significantly protective effects, K222 or D/S 146, which were not detected in the samples of the analyzed Indonesian breeds, were found in other Asian or Eurasian populations at very low frequencies, e.g., in some Chinese, Japanese, Korean, Pakistani, and Turkish breeds. Only in the Turkish Halep breed, the S146 frequency was higher with 28% (Hussain et al. 2011; Zhou et al. 2013; Hassan et al. 2016; Meydan et al. 2017; Kim et al. 2019; Akis et al. 2020).
Due to insufficient passive surveillance systems, the actual prevalence of scrapie remains unknown in many countries (Curcio et al. 2016); this is also the case in Indonesia, where no scrapie cases have been reported until now. Therefore, their apparent genetic susceptibility based on PRNP polymorphisms seems not to pose an acute risk to the goat breeds studied. However, based on current knowledge, it has to be concluded that these breeds are no relevant resource for resistance breeding programs in the case this might be necessary in the future. However, in order to represent each region of the Indonesian archipelago, research on Lakor, Marica, Bligon, Muara, and Kosta goat breeds, other Indonesian goat breeds which were not included in this study, should be done, using additional samples and other sampling locations.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Acín, C., Martín-Burriel, I., Monleón, E., Lyahyai, J., Pitarch, J.L., Serrano, C., Monzòn, M., Zaragoza, P., Badiola, J.J., 2013. Prion protein gene variability in Spanish goats inference through susceptibility to classical scrapie strains and pathogenic distribution of peripheral PrPsc. PloS One, 8(4): e61118.
Acutis, P.L., Bossers, A., Priem, J., Riina, M.V., Peletto, S., Mazza, M., Casalone, C., Forloni, G., Ru, G., Caramelli, M., 2006. Identification of prion protein gene polymorphisms in goats from Italian scrapie outbreaks. Journal of General Virology, 87, 1029–1033.
Acutis, P.L., Colussi, S., Santagada, G., Laurenza, C., Maniaci, M.G., Riina, M.V., Peletto, S., Goldmann, W., Bossers, A., Caramelli, M., Cristoferi, I., Maione, S., Sacchi, P., Rasero, R., 2008. Genetic variability of the PRNP gene in goat breeds from Northern and Southern Italy. Journal of Applied Microbiology, 104, 1782–1789.
Akis, I., Oztabak, K., Atmaca, G., Gursel, F. E., Ates, A., Yardibi, H., Gurgoze, S., Durak, M. H., Erez, I., Un, C., 2020. PRNP gene polymorphisms in main indigenous Turkish goat breeds. Tropical animal health and production, 52, 793-802.
Babar, M.E., Nawaz, M., Nasim, A., Imran, M., Jabeen, R., Chatha, S.A., Haq, A.U., Nawaz, A., Mustafa, H., Nadeem, A., 2008. Prion protein genotypes in Pakistani goats. Asian Australasian Journal of Animal Sciences, 21, 936–940.
Barillet, F., Mariat, D., Amigues, Y., Faugeras, R., Caillat, H., Moazami-Goudarzi, K., Rupp, R., Babilliot, J.M., Lacroux, C., Lugan, S., Schelcher, F., Chartier, C., Corbière, F., Andréoletti, O., Perrin-Chauvineau, C., 2009. Identification of seven haplotypes of the caprine PrP gene at codons 127, 142, 154, 211, 222 and 240 in French Alpine and Saanen breeds and their association with classical scrapie. Journal of General Virology, 90(Pt 3):769-776.
Baylis M., Chihota C., Stevenson E., Goldmann W., Smith A., Sivam K., Tongue S., Gravenor M.B., 2004. Risk of scrapie in British sheep of different prion protein genotype. Journal of General Virology, 85, 2735-2740.
Benestad, S. L., Sarradin, P., Thu, B., Schönheit, J., Tranulis, M. A., Bratberg, B., 2003. Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. Veterinary Record, 153, 202-208.
Billinis, C., Panagioidis, H.C., Psychas, V., Argyroudis, S., Nicolaou, A., Leontides, S., Papadopoulos, O., Sklaviadis, T., 2002. Prion protein gene polymorphisms in natural goat scrapie. Journal of General Virology, 83, 713–721.
Chelle, P. L., 1942. A case of trembling in the goat. Academie Veterinaire de France Bulletin, 15, 294–295.
Cinar, M. U., Schneider, D. A., Waldron, D. F., O’Rourke, K. I., White, S. N., 2018. Goats singly heterozygous for PRNP S146 or K222 orally inoculated with classical scrapie at birth show no disease at ages well beyond 6 years. The Veterinary Journal, 233, 19-24.
Curcio, L., Sebastiani, C., Di Lorenzo, P., Lasagna, E., Biagetti, M., 2016. Review: A review on classical and atypical scrapie in caprine: prion protein gene polymorphisms and their role in the disease. Animal, 10, 1585-93.
Dawson, M., Hoinville, L. J., Hosie, B. D., Hunter, N., 1998. Guidance on the use of PrP genotyping as an aid to the control of clinical scrapie. Veterinary Record, 142, 623-625.
Dubois, M. A., Sabatier, P., Durand, B., Calavas, D., Ducrot, C., Chalvet-Monfray, K., 2002. Multiplicative genetic effects in scrapie disease susceptibility. Comptes Rendus Biologies, 325(5), 565-570.
EFSA, 2012. Scientific and technical assistance on the provisional results of the study on genetic resistance to classical scrapie in goats in Cyprus. EFSA Journal, 10(11), 2972
EFSA Panel on Biological Hazards (BIOHAZ), 2014. Scientific opinion on the scrapie situation in the EU after 10 years of monitoring and control in sheep and goats. EFSA Journal, 12(7), 3781.
EFSA Panel on Biological Hazards (BIOHAZ), 2017. Scientific opinion on genetic resistance to transmissible spongiform encephalopathies (TSE) in goats. EFSA Journal, 15(8), 4962.
Eiden, M., Soto, E. O., Mettenleiter, T. C., Groschup, M. H., 2011. Effects of polymorphisms in ovine and caprine prion protein alleles on cell-free conversion. Veterinary Research, 42, 1-7.
Fragkiadaki, E.G., Vaccari, G., Ekateriniadou, L.V., Agrimi, U., Giadinis, N.D., Chiappini, B., et al., 2011. PRNP genetic variability and molecular typing of natural goat scrapie isolates in a high number of infected flocks. Veterinary Research, 42, 104.
Goldmann, W., 2008. PrP genetics in ruminant transmissible spongiform encephalopathies. Veterinary Research, 39(4), 1-14.
Goldmann, W., Martin, T., Foster, J., Hughes, S., Smith, G., Hughes, K., Dawson, M., Hunter, N., 1996. Novel polymorphisms in the caprine PrP gene: a codon 142 mutation associated with scrapie incubation period. Journal of General Virology, 77, 2885–2891.
Goldmann, W., Chong, A., Foster, J., Hope, J., Hunter, N., 1998. The shortest known prion protein gene allele occurs in goats, has only three octapeptide repeats and is non-pathogenic. Journal of General Virology, 79, 3173–3176.
Goldmann, W., Ryan, K., Stewart, P., Parnham, D., Xicohtencatl, R., Fernandez, N., Saunders, G., Windl, O., González, L., Bossers, A., Foster, J., 2011. Caprine prion gene polymorphisms are associated with decreased incidence of classical scrapie in goat herds in the United Kingdom. Veterinary Research, 42(1):110.
Goldmann, W., Marier, E., Stewart, P., Konold, T., Street, S., Langeveld, J., Windl, O., Ortiz-Pelaez, A., 2016. Prion protein genotype survey confirms low frequency of scrapie-resistant K222 allele in British goat herds. Veterinary Record, 178, 168–174.
Hassan, M.F., Khan, S.H., Babar, M.E., Yang, L., Ali, T., Khan, J.M., Shah, S.Z., Zhou, X., Hussain, T., Zhu, T., Hussain, T., Zhao, D., 2016. Polymorphism analysis of prion protein gene in 11 Pakistani goat breeds. Prion, 10(4), 290-304.
Hussain, A., Babar, M. E., Imran, M., Haq, I. U., Javed, M. M., 2011. Detection of four novel polymorphisms in PrP gene of Pakistani sheep (Damani and Hashtnagri) and goats (Kamori and Local Hairy) breeds. Virology Journal, 8(1), 1-5.
Kim, S. K., Kim, Y. C., Won, S. Y., Jeong, B. H., 2019. Potential scrapie-associated polymorphisms of the prion protein gene (PRNP) in Korean native black goats. Scientific Reports, 9(1), 15293.
Kipanyula, M. J., Chuma, I. S., Brundtland, E., Bardsen, K., Msalya, G., Kifaro, G. C., Ulvund, M. J., 2014. Prion protein (PrP) gene polymorphisms in Small East African and Norwegian white goats. Small Ruminant Research, 121, 200–206.
Kurosaki, Y., Ishiguro, N., Horiuchi, M., Shinagawa, M., 2005. Polymorphisms of caprine PrP gene detected in Japan. Journal of veterinary medical science, 67(3), 321-323.
Lühken, G., Buschmann, A., Brandt, H., Eiden, M., Groschup, M. H., Erhardt, G., 2007. Epidemiological and genetical differences between classical and atypical scrapie cases. Veterinary Research, 38(1), 65-80
Meydan, H., Pehlivan, E., Özkan, M. M., Yildiz, M. A., Goldmann, W., 2017. Prion protein gene polymorphisms in Turkish native goat breeds. Journal of Genetics, 96(2), 299-305.
Pakpahan, S., Artama, W. T., Widayanti, R., Suparta, I. G., 2016. Molecular phylogenetic of Hutan Sumatera goat (Sumatran Serow) and domestic goat (Capra hircus) in Indonesia based on analysis mitochondrial cytochrome b gene. Asian Journal of Animal Veterinary Advances, 11(6), 331-340.
Papasavva-Stylianou, P., Windl, O., Saunders, G., Mavrikiou, P., Toumazos, P., Kakoyiannis, C., 2011. PrP gene polymorphisms in Cyprus goats and their association with resistance or susceptibility to natural scrapie. The Veterinary Journal, 187(2), 245-250.
Prusiner, S. B., DeArmond, S. J., 1994. Prion diseases and neurodegeneration. Annu Rev Neurosci, 17, 311–339.
Stahl, N., Baldwin, M. A., Burlingame, A. L., Prusiner, S. B., 1990. Identification of glycoinositol phospholipid-linked and truncated forms of the scrapie prion protein. Biochemistry, 29(38), 8879-8884.
Stöckel, J., Safar, J., Wallace, A. C., Cohen, F. E., Prusiner, S. B., 1998. Prion protein selectively binds copper (II) ions. Biochemistry, 37, 7185–7193.
Vaccari, G., Di Bari, M.A., Morelli, L., Nonno, R., Chiappini, B., Antonucci, G., Marcon, S., Esposito, E., Fazzi, P., Palazzini, N., Troiano, P., Petrella, A., Di Guardo, G., Agrimi, U., 2006. Identification of an allelic variant of the goat PrP gene associated with resistance to scrapie. Journal of General Virology, 87, 1395–1402.
Vaccari, G., Panayiotidis, C.H., Acin, C., Peletto, S., Barillet, F., Acutis, P.L., Bossers, A., Langeveld, J., van Keulen, L., Sclaviadis, T., Badiola, J.J., Andreoletti, O., Groschup, M.H., Agrimi, U., Foster, J., Goldman, W., 2009. State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Veterinary Research, 40, 1- 18.
Vitezica, Z. G., Elsen, J. M., Rupp, R., Díaz, C., 2005. Using genotype probabilities in survival analysis: a scrapie case. Genetics Selection Evolution, 37(5), 1-13.
White, S., Herrmann-Hoesing, L., O'rourke, K., Waldron, D., Rowe, J., Alverson, J., 2008. Prion gene (PRNP) haplotype variation in United States goat breeds (Open Access publication). Genetics Selection Evolution, 40(5), 553-561.
Windig, J. J., Hoving, R. A. H., Priem, J., Bossers, A., Van Keulen, L. J. M., Langeveld, J. P. M., 2016. Variation in the prion protein sequence in Dutch goat breeds. Journal of Animal Breeding and Genetics, 133(5), 366-374.
Zhou, R., Li, X., Xi, J., Li, L., Zhang, Z., Zhao, Z., 2013. Genetic variability of PRNP in Chinese indigenous goats. Biochemical Genetics, 51(3), 211-222.
Acknowledgements
The authors are thankful to the Direktorat Penelitian dan Tim Peningkatan Reputasi UGM menuju World Class University-Kantor Jaminan Mutu UGM, for funding this study in the post-doctoral program (grant no. 6162/UN1/DITLIT/DIT-LIT/PT/2021). We thank the farmers and people who helped in the collection of samples and the Indonesian Livestock Service Office for supporting this research. The authors would also like to thank Dela Ayu Lestari for helping with sample collection, as well as Carina Crispens and Stephanie Steitz for technical assistance.
Funding
Open Access funding enabled and organized by Projekt DEAL. SP was financially supported by the Direktorat Penelitian dan Tim Peningkatan Reputasi UGM menuju World Class University-Kantor Jaminan Mutu UGM in a post-doctoral program (grant no. 6162/UN1/DITLIT/DIT-LIT/PT/2021).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Material preparation, data collection, and analysis were performed by SP and GL. The first draft of the manuscript was written by SP, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the Animal Ethics Committee for using Animal and Scientific Procedures in the Faculty of Veterinary Medicine, Universitas Gadjah Mada, Indonesia, nr.003/EC-FKH/Eks./2022.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pakpahan, S., Widayanti, R., Artama, W.T. et al. Genetic variability of the prion protein gene in Indonesian goat breeds. Trop Anim Health Prod 55, 87 (2023). https://doi.org/10.1007/s11250-023-03486-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11250-023-03486-7