Abstract
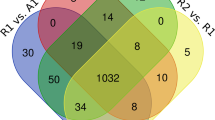
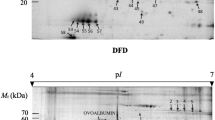
The biosafety of fat-1 transgenic cattle has been a focus of our studies since the first fat-1 transgenic cow was born. In this study, we used tandem mass tag labeling, TiO2 enrichment, and nanoscale liquid chromatography coupled with tandem mass spectrometry (nanol LC–MS/MS) to compare proteomic and phosphoproteomic profiling analyses of muscle between fat-1 transgenic cows and wild-type cows. A total of 1555 proteins and 900 phosphorylation sites in 159 phosphoproteins were identified in the profiling assessments, but only four differentially expressed proteins and nine differentially expressed phosphopeptides were detected in fat-1 transgenic cows relative to wild-type cows. Bioinformatics analyses showed that all of the identified proteins and phosphoproteins were mainly related to the metabolic processes of three major nutrients: carbohydrates, lipids, and proteins. All of these differentially expressed proteins might take part in DNA recombination, repair, and regulation of the immune system. In conclusion, most of the identified proteins and phosphoproteins exhibited few changes. Our results provide new insights into the biosafety of fat-1 transgenic cattle.



Similar content being viewed by others
References
Andruchov O, Andruchova O, Wang Y, Galler S (2006) Dependence of cross-bridge kinetics on myosin light chain isoforms in rabbit and rat skeletal muscle fibres. J Physiol 571:231–242. doi:10.1113/jphysiol.2005.099770
Beningo KA, Hamao K, Dembo M, Wang YL, Hosoya H (2006) Traction forces of fibroblasts are regulated by the Rho-dependent kinase but not by the myosin light chain kinase. Arch Biochem Biophys 456:224–231. doi:10.1016/j.abb.2006.09.025
Bhattacharya A, Lawrence RA, Krishnan A, Zaman K, Sun D, Fernandes G (2003) Effect of dietary n-3 and n-6 oils with and without food restriction on activity of antioxidant enzymes and lipid peroxidation in livers of cyclophosphamide treated autoimmune-prone NZB/W female mice. J Am Coll Nutr 22:388–399
Carlo C, Pura B, Magaly R, Marino D (2016) Differential effects of contractile potentiators on action potential-induced Ca2+ transients of frog and mouse skeletal muscle fibres. J Muscle Res Cell Motil. doi:10.1007/s10974-016-9455-3
Celeste A et al (2002) Genomic instability in mice lacking histone H2AX. Science 296:922–927. doi:10.1126/science.1069398
Cha IS et al (2012) Kidney proteome responses in the teleost fish Paralichthys olivaceus indicate a putative immune response against Streptococcus parauberis. J Proteom 75:5166–5175. doi:10.1016/j.jprot.2012.05.046
Chardin P (2003) GTPase regulation: getting aRnd Rock and Rho inhibition. Curr Biol 13:R702–R704
Clarke SE, Kang JX, Ma DW (2014) The iFat1 transgene permits conditional endogenous n-3 PUFA enrichment both in vitro and in vivo. Transgenic Res 23:489–501. doi:10.1007/s11248-014-9788-x
Cleland LG, James MJ, Proudman SM (2003) The role of fish oils in the treatment of rheumatoid arthritis. Drugs 63:845–853
Gill I, Valivety R (1997) Polyunsaturated fatty acids, part 1: occurrence, biological activities and applications. Trends Biotechnol 15:401–409. doi:10.1016/S0167-7799(97)01076-7
Gonzalez-Romero R, Rivera-Casas C, Frehlick LJ, Mendez J, Ausio J, Eirin-Lopez JM (2012) Histone H2A (H2A.X and H2A.Z) variants in molluscs: molecular characterization and potential implications for chromatin dynamics. PLoS ONE 7:e30006. doi:10.1371/journal.pone.0030006
Guo J, He K, Bai S, Zhang T, Liu Y, Wang F, Wang Z (2016) Effects of transgenic cry1Ie maize on non-lepidopteran pest abundance, diversity and community composition. Transgenic Res. doi:10.1007/s11248-016-9968-y
Harari-Steinberg O, Cantera R, Denti S, Bianchi E, Oron E, Segal D, Chamovitz DA (2007) COP9 signalosome subunit 5 (CSN5/Jab1) regulates the development of the Drosophila immune system: effects on Cactus, Dorsal and hematopoiesis. Genes Cells 12:183–195. doi:10.1111/j.1365-2443.2007.01049.x
Himpens B, Matthijs G, Somlyo AV, Butler TM, Somlyo AP (1988) Cytoplasmic free calcium, myosin light chain phosphorylation, and force in phasic and tonic smooth muscle. J Gen Physiol 92:713–729
Hollis GF, Evans RJ, Stafford-Hollis JM, Korsmeyer SJ, McKearn JP (1989) Immunoglobulin lambda light-chain-related genes 14.1 and 16.1 are expressed in pre-B cells and may encode the human immunoglobulin omega light-chain protein. Proc Natl Acad Sci USA 86:5552–5556
Hu FB et al (2002) Fish and omega-3 fatty acid intake and risk of coronary heart disease in women. JAMA 287:1815–1821
Humphries JM et al (2014) Identification and validation of novel candidate protein biomarkers for the detection of human gastric cancer. Biochem Biophys Acta 1844:1051–1058. doi:10.1016/j.bbapap.2014.01.018
Ikebe R, Reardon S, Mitsui T, Ikebe M (1999) Role of the N-terminal region of the regulatory light chain in the dephosphorylation of myosin by myosin light chain phosphatase. J Biol Chem 274:30122–30126
Iso H et al (2001) Intake of fish and omega-3 fatty acids and risk of stroke in women. JAMA 285:304–312
Kang JX, Wang J, Wu L, Kang ZB (2004) Transgenic mice: fat-1 mice convert n-6 to n-3 fatty acids. Nature 427:504. doi:10.1038/427504a
Kwon JH, Lee YM, An CS (2000) CDNA cloning of chloroplast omega-3 fatty acid desaturase from Capsicum annuum and its expression upon wounding. Mol Cells 10:493–497
Lai L et al (2006) Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat Biotechnol 24:435–436. doi:10.1038/nbt1198
Larsson SC, Kumlin M, Ingelman-Sundberg M, Wolk A (2004) Dietary long-chain n-3 fatty acids for the prevention of cancer: a review of potential mechanisms. Am J Clin Nutr 79:935–945
Lema MA, Burachik M (2009) Safety assessment of food products from r-DNA animals. Comp Immunol Microbiol Infect Dis 32:163–189. doi:10.1016/j.cimid.2007.11.007
Liu Y, Schiff M, Serino G, Deng XW, Dinesh-Kumar SP (2002) Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14:1483–1496
Liu X, Bai C, Ding X, Wei Z, Guo H, Li G (2015) Microarray analysis of the gene expression profile and lipid metabolism in fat-1 transgenic cattle. PLoS ONE 10:e0138874. doi:10.1371/journal.pone.0138874
Lu ZQ et al (2016) Maternal dietary linoleic acid supplementation promotes muscle fibre type transformation in suckling piglets. J Anim Physiol Anim Nutr. doi:10.1111/jpn.12626
Marancik DP, Fast MD, Camus AC (2013) Proteomic characterization of the acute-phase response of yellow stingrays Urobatis jamaicensis after injection with a Vibrio anguillarum-ordalii bacterin. Fish Shellfish Immunol 34:1383–1389. doi:10.1016/j.fsi.2013.02.024
Menon S, Chi H, Zhang H, Deng XW, Flavell RA, Wei N (2007) COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat Immunol 8:1236–1245. doi:10.1038/ni1514
Meyer C, Dostou JM, Welle SL, Gerich JE (2002) Role of human liver, kidney, and skeletal muscle in postprandial glucose homeostasis. Am J Physiol Endocrinol Metab 282:E419–E427. doi:10.1152/ajpendo.00032.2001
Minami A, Ishimura N, Sakamoto S, Takishita E, Mawatari K, Okada K, Nakaya Y (2002) Effect of eicosapentaenoic acid ethyl ester v. oleic acid-rich safflower oil on insulin resistance in type 2 diabetic model rats with hypertriacylglycerolaemi. Br J Nutr 87:157–162. doi:10.1079/BJN2001496
Nakamura A et al (2008) Role of non-kinase activity of myosin light-chain kinase in regulating smooth muscle contraction, a review dedicated to Dr. Setsuro Ebashi. Biochem Biophys Res Commun 369:135–143. doi:10.1016/j.bbrc.2007.11.096
Nemets B, Stahl Z, Belmaker RH (2002) Addition of omega-3 fatty acid to maintenance medication treatment for recurrent unipolar depressive disorder. Am J Psychiatry 159:477–479. doi:10.1176/appi.ajp.159.3.477
Peet M, Brind J, Ramchand CN, Shah S, Vankar GK (2001) Two double-blind placebo-controlled pilot studies of eicosapentaenoic acid in the treatment of schizophrenia. Schizophr Res 49:243–251
Pratt VC, Watanabe S, Bruera E, Mackey J, Clandinin MT, Baracos VE, Field CJ (2002) Plasma and neutrophil fatty acid composition in advanced cancer patients and response to fish oil supplementation. Br J Cancer 87:1370–1378. doi:10.1038/sj.bjc.6600659
Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W (2002) Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev 12:162–169
Redon CE et al (2010) Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin Cancer Res 16:4532–4542. doi:10.1158/1078-0432.CCR-10-0523
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868
Ross PL et al (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteom 3:1154–1169. doi:10.1074/mcp.M400129-MCP200
Russo JM et al (2005) Distinct temporal-spatial roles for rho kinase and myosin light chain kinase in epithelial purse-string wound closure. Gastroenterology 128:987–1001
Schulze WX, Usadel B (2010) Quantitation in mass-spectrometry-based proteomics. Annu Rev Plant Biol 61:491–516. doi:10.1146/annurev-arplant-042809-112132
Schwartz SA, Hernandez A, Mark Evers B (1999) The role of NF-kappaB/IkappaB proteins in cancer: implications for novel treatment strategies. Surg Oncol 8:143–153
Spychalla JP, Kinney AJ, Browse J (1997) Identification of an animal omega-3 fatty acid desaturase by heterologous expression in Arabidopsis. Proc Natl Acad Sci USA 94:1142–1147
Sweeney HL, Bowman BF, Stull JT (1993) Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am J Physiol 264:C1085–C1095
Tang M et al (2011) Safety assessment of sFat-1 transgenic pigs by detecting their co-habitant microbe in intestinal tract. Transgenic Res 20:749–758. doi:10.1007/s11248-010-9457-7
Tang M et al (2014) Functional and safety evaluation of transgenic pork rich in omega-3 fatty acids. Transgenic Res 23:557–571. doi:10.1007/s11248-014-9796-x
Taus T, Kocher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K (2011) Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res 10:5354–5362. doi:10.1021/pr200611n
Tomoda K, Kubota Y, Kato J (1999) Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398:160–165. doi:10.1038/18230
Tsai WS, Nagawa H, Muto T (1997) Differential effects of polyunsaturated fatty acids on chemosensitivity of NIH3T3 cells and its transformants. Int J Cancer 70:357–361
Tzolovsky G, Millo H, Pathirana S, Wood T, Bownes M (2002) Identification and phylogenetic analysis of Drosophila melanogaster myosins. Mol Biol Evol 19:1041–1052
Van Reenen CG, Meuwissen TH, Hopster H, Oldenbroek K, Kruip TH, Blokhuis HJ (2001) Transgenesis may affect farm animal welfare: a case for systematic risk assessment. J Anim Sci 79:1763–1779
Vidal N, Barbosa H, Jacob S, Arruda M (2015) Comparative study of transgenic and non-transgenic maize (Zea mays) flours commercialized in Brazil, focussing on proteomic analyses. Food Chem 180:288–294. doi:10.1016/j.foodchem.2015.02.051
Wang ZK, Wang J, Liu J, Ying SH, Peng XJ, Feng MG (2016) Proteomic and phosphoproteomic insights into a signaling hub role for Cdc14 in asexual development and multiple stress responses in Beauveria bassiana. PLoS ONE 11:e0153007. doi:10.1371/journal.pone.0153007
Wierer M, Mann M (2016) Proteomics to study DNA-bound and chromatin-associated gene regulatory complexes. Hum Mol Genet. doi:10.1093/hmg/ddw208
Wolfe RR (2006) The underappreciated role of muscle in health and disease. Am J Clin Nutr 84:475–482
Wu X et al (2012) Production of cloned transgenic cow expressing omega-3 fatty acids. Transgenic Res 21:537–543. doi:10.1007/s11248-011-9554-2
Xu J, Zhao J, Wang J, Zhao Y, Zhang L, Chu M, Li N (2011) Molecular-based environmental risk assessment of three varieties of genetically engineered cows. Transgenic Res 20:1043–1054. doi:10.1007/s11248-010-9477-3
Yam D, Peled A, Shinitzky M (2001) Suppression of tumor growth and metastasis by dietary fish oil combined with vitamins E and C and cisplatin. Cancer Chemother Pharmacol 47:34–40. doi:10.1007/s002800000205
Yamanouchi K (2007) Regulatory considerations on transgenic livestock in Japan in relation to the Cartagena protocol. Theriogenology 67:185–187. doi:10.1016/j.theriogenology.2006.09.009
Yang X et al (2002) The COP9 signalosome inhibits p27(kip1) degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr Biol 12:667–672
Yang Y, Qiang X, Owsiany K, Zhang S, Thannhauser TW, Li L (2011) Evaluation of different multidimensional LC–MS/MS pipelines for isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis of potato tubers in response to cold storage. J Proteome Res 10:4647–4660. doi:10.1021/pr200455s
Yang QS et al (2012) Quantitative proteomic analysis reveals that antioxidation mechanisms contribute to cold tolerance in plantain (Musa paradisiaca L.; ABB Group) seedlings. Mol Cell Proteom 11:1853–1869. doi:10.1074/mcp.M112.022079
Zhu G, Chen H, Wu X, Zhou Y, Lu J, Chen H, Deng J (2008) A modified n-3 fatty acid desaturase gene from Caenorhabditis briggsae produced high proportion of DHA and DPA in transgenic mice. Transgenic Res 17:717–725. doi:10.1007/s11248-008-9171-x
Zou S, Huang K, Xu W, Luo Y, He X (2016) Safety assessment of lepidopteran insect-protected transgenic rice with cry2A* gene. Transgenic Res 25:163–172. doi:10.1007/s11248-015-9920-6
Acknowledgements
This work was supported by the National Transgenic Animal Program (Grant Nos. 2014ZX08007-002, 2016ZX08007002-004) of China. This work was accomplished in the Tianjin International Joint Research and Development Center. We thank The Key Laboratory of Mammalian Reproductive Biology and Biotechnology of the Ministry of Education, Inner Mongolia University, for proving us with experimental materials. We also want to thank the Proteomic and MS Facility of Cornell University and the team of Professor Sheng Zhang for generating the mass spectrometry data and NIH SIG 1S10 OD017992-01 grant support for the Orbitrap Fusion mass spectrometer. We are grateful to the help of Agricultural animal breeding and product testing Biotechnology Research Center of Tianjin Agricultural University.
Author information
Authors and Affiliations
Contributions
Author contributions
HG, XFL, GPL and XBX conceived and designed the experiments. XBX and SPY performed most of the experiments, and XBX performed the data analyses and wrote the manuscript. CFJ and SPY contributed significantly to performed animal samples preparation. XL, XBD and XFL contributed significantly to revise the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that no conflict of interest exists in the manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xin, X., Liu, X., Li, X. et al. Comparative muscle proteomics/phosphoproteomics analysis provides new insight for the biosafety evaluation of fat-1 transgenic cattle. Transgenic Res 26, 625–638 (2017). https://doi.org/10.1007/s11248-017-0032-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-017-0032-3