Abstract
The maize Emb5 is an abscisic acid–responsive gene which is specifically expressed in the late embryo during seed maturity. To further dissect and identify the elements specific for its embryo expression pattern, we investigated the activity of the − 1653 bp upstream of the “full-length” promoter region of this gene in transgenic Arabidopsis plants. We first confirmed that the “full-length” promoter could indeed drive the expression of β-glucuronidase reporter gene (GUS) in the transgenic Arabidopsis seed embryo. Subsequently, DNA fragments of ~ 500 bp in length were generated after a series of progressive deletions from positions − 1653 bp to − 1 bp relative to the transcriptional initiation site. These fragments were fused with GUS and introduced into Arabidopsis. Measurement of the GUS activity in the immature seeds isolated from the transgenic plants revealed that the region between positions − 523 bp and − 1 bp, namely ProEm-D, is absolutely required and sufficient for the temporal and embryo-specific expression of GUS with an activity comparable with the full-length Emb5 promoter in Arabidopsis. Therefore, our results clearly demonstrated that the 523 bp ProEm-D can replace the − 1653 bp Emb5 promoter to drive embryo-specific expression in Arabidopsis seed. Because of its small size and strong embryo-specific activity, it could become the promoter of choice in metabolic pathway engineering to transfer multiple genes for the production of valuable pharmaceutical products in seeds, such as polyunsaturated fatty acids found in fish oils, or pro-vitamin A where at least three transgenes are required to assemble the entire metabolic pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Using corn seeds to produce recombinant proteins for human consumption has many advantages, including low probability of contamination by human and animal endotoxins, relatively high protein yield, and low production cost. In addition, as a eukaryote, corn presents a clear advantage compared with the E. coli expression platform in order to express genes originated from eukaryotic organisms due to codon compatibility as well as to express those proteins that require post-translational modification (Pan et el. 2015). To achieve this, embryo-specific promoters are required to drive the expression of exogenous genes and production of certain proteins in seeds of corn. However, only very few tissue-specific promoters, especially those embryo-specific, are available at present, hence limiting the utility of corn seeds in the biotechnology industry.
Strong constitutive promoters such as the Cauliflower Mosaic Virus 35S (CaMV 35S) promoter and maize ubiquitin promoter have been routinely and widely used to drive gene expression for plant research and biotechnology (Battraw and Hall 1990; Christensen et al. 1992; Saidi et al. 2009; Li et al. 2015). However, the expression of target gene(s) in the desired tissue or/and developmental stages cannot be controlled due to their constitutive nature of expression activities (Drakakaki et al. 2000). This could lead to the persistent and high-level expression of a foreign protein(s) in all tissues throughout the growth and development stages, which may have some negative effect on the host plants (Cheon et al. 2004; Lescto et al. 2002; Yamori 2013). In contrast, expression of a gene-of-interest controlled by tissue- or organ-specific promoters could often eliminate/reduce these adverse effects. Therefore, it is desirable to isolate such promoters for basic research as well as plant biotechnology to produce valuable proteins or to create specific traits in crop plants (Naoumkina et al. 2008; Naoumkina and Dixon 2011).
To this end, research on the isolation and characterization of plant seed–specific expression promoters has attracted a great deal of attention; hence, much progress has been made in recent years. For example, by expressing the promoter region of the peanut 8A4R19G1 gene in the model plants Arabidopsis and tobacco, this groundnut seed promoter (GSP) showed seed-specific activity (Sunkara et al. 2014). Similarly, the Napin promoter of Brassica napus has been confirmed to drive seed-specific expression of GUS, and it has been widely used to express many genes in dicotyledonous plants (Sohrabi et al. 2015). A few seed-specific promoters from maize have also been isolated recently. One of them is the globulin-1 promoter where the specific expression of foreign genes in the maize embryos was reported (Requesens et al. 2019). However, the amount of protein produced is very low despite the strong activity of this promoter because the embryo only accounts for 10% of the total seed weight. To increase the protein quantity in the seeds, an endosperm-specific promoter was used. This was proved to be a much better promoter of choice because the large proportion of the corn seed is made up of endosperm. However, corn endosperm contains mainly starch, while the embryo is rich in linolenic acid and other unsaturated fatty acids that can be manipulated for the production of specific corn germ oils, for example. In addition, seeds of many oilseed crops, such as oilseed rape and other Brassica crops (close relatives of Arabidopsis), groundnuts, and beans are made up mainly by embryos (> 90%) because the growing embryo absorbs all the endosperm so at maturity no endosperm is evident. Therefore, expression of recombinant proteins in seeds driven by embryo-specific promoters in these oilseed crops presents clear advantages over endosperm-driven expression.
To determine the activity of the 635 bp bidirectional promoter proZmBD1 of the two defense genes Def1 and Def2 in maize, GUS and GFP were fused to proZmBD1. Both transient and stable expressions were carried out, and it was demonstrated that GUS and GFP could be specifically expressed in the transgenic maize embryos (Liu et al. 2016). Pan et al. (2015) studied the functionality of the 1053 bp long promoter sequence of the millet pF128 gene in transgenic Arabidopsis, millet, and maize plants. The specific expression of GUS was detected in the seeds of the transgenic lines of all these three plant species. Interestingly, they also noted that the activity of this millet seed-specific promoter was much higher than that of the routinely used constitutive CaMV35S promoter and the maize seed–specific promoter p19Z.
In this study, we carried out an in-depth and systematic analysis of the structural elements of a previously isolated embryo-specific promoter of the Emb5 gene from maize (Kriz et al. 2006, US Patent No. US7,078,234 B2). The function of the cis-acting elements and the core functional regions associated with the embryo-specific activity of this promoter were identified by deletion experiments. As a result, we identified a much shorter sequence of 523 bp within the core region of this promoter that still retains the strong and embryo-specific activity. This newly isolated promoter sequence of maize may provide more potential in basic research as well as biotechnology in maize as well as other crop plants, particularly in the assembly of complex metabolic and/or signaling pathways where expression of multiple genes is required.
Materials and Methods
Cloning of the Full-Length Promoter of the Emb5 Gene
The genomic DNA was extracted by the CTAB method from young leaves of seedlings of maize inbred line Qi319 (Aboul-Maaty and Oraby 2019). The full-length sequence of the Emb5 promoter (Kriz et al. 2006) was amplified by gradient PCR with high-fidelity DNA polymerase (Takara) using the Emb5 promoter specific primers Emb5L and Emb5R (Table 1S).
Construction of proEmb5:GUS Plant Expression Vector and the Acquisition of Transgenic Arabidopsis Plants
The PCR product of proEmb5 was ligated into the cloning vector pMD18-T Simple (Tanaka, Japan) and sequenced. To construct the plant expression vector, the proEmb5 was released from pMD18-T by double-digestion with EcoRI and PstI, purified and ligated to the same sites of the plant expression vector pBI121, replacing the original CaMV35S (35S) promoter. This resulted in the production of pBI121-proEmb5:GUS. For negative control, the 54 bp of 35S minimal promoter (Sohrabi et al. 2015), which is not expressed in a plant, was used. pBI121, which has the 35S:GUS expression cassette, was used as the positive control in the subsequent Arabidopsis transformation studies.
To make the seven 5′-end deletion constructs containing proEmbA-G (Fig. 2a), PCR amplification was carried out with the individual corresponding primer pairs (Suppl Table 1S), using the pMD18-T Simple-ProEmb5 generated as above as the template. After confirmation by colony PCR and sequencing, the individual DNA fragments were subsequently ligated to pBI121 to generate pBI121-proEmbA-G-GUS constructs. In addition, the individual conserved region of the ProEmb5 was added by overlapping PCR to upstream of the 54 bp 35S minimal promoter (Sohrabi et al. 2015). These fragments were subsequently ligated to the pBI121 vector, replacing of the original 35S promoter to result in the production of a series of GUS expression constructs as indicated in Fig. 6.
To study the regulatory regions within the fragments En (− 1653 to − 1143, positive regulatory region 1) and Dn (− 523 to − 331, positive regulatory region 2), PCR was carried out with primer pairs targeting these specific regions (Suppl Table 1S). The resulting products were cloned into pMD18-T simple vector and again sequenced. Inserts with correct sequences were double digested with EcoRI and PstI and ligated to the same sites of the linearized pMD18-T simple-35S minimal promoter to obtain pMD-En-35Smp and pMD-Dn-35Smp.
To make the ProEn-Dn-35Smp:GUS and ProDn-Dn-35Smp:GUS constructs, En and Dn were first released by digesting pMD-En-35Smp and pMD-Dn-35Smp with HindШ and EcoRI. These were ligated to the upstream of Dn in the pMD-Dn-35Smp, generating pMD-En-Dn-35Smp:GUS and pMD-Dn-Dn-35Smp:GUS, respectively. After sequencing confirmation of the identity of the En-Dn and Dn-Dn, they were excised with HindШ and BamHI and ligated to the same sites of pBI121, again replacing the original 35S promoter to result in the production of proEn-Dn:GUS and proDn-Dn:GUS constructs for plant transformation.
Plant Transformation
Wild-type Arabidopsis plants of ecotype Col-0 were transformed by the floral dipping method (Clough and Bent 1998). Seeds were collected, and transgenic plants were screened on ½ MS medium containing 40 μg/ml kanamycin. After 10 days, surviving seedlings were transferred to soil and grown in an environmentally controlled growth room (16 h light/8 h darkness at 21 °C) until maturity. PCR was carried out to confirm the presence of individual DNA fragments (data not shown). Transgenic plants containing a single copy (observed by 1:3 segregation rate) of the transgenes were grown for 3 generations in order to obtain homozygous plants for subsequent GUS expression analysis as detailed below.
GUS Histochemical Staining
Arabidopsis plant tissues from 4 to 6 independent transgenic homozygous lines were isolated and placed in cold 90% acetone for 10 min. They were then rinsed with 50 mM phosphate buffer (pH 7.0) before adding the staining solution (1 mM X-Gluc, 50 mM sodium phosphate, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6) and incubated at 37 °C in the dark for 16–24 h. All positively stained tissues were incubated in the solution for 16 h, while tissues that did not show significant staining were incubated for up to 24 h (Jefferson 1987). To stop the reaction, 70% ethanol was added. After 12–48 h, the 70% ethanol was replaced with 95% ethanol, and the seedlings were stored at room temperature, observed, and photographed.
Quantitative Fluorometric GUS Assays
The same tissues used for the GUS staining assays above were ground in the GUS extraction buffer (50 mmol Na3PO4(PH 7.0), 10 mmol EDTA, 0.1% Triton X-100, 0.1% Sarcosyl, 10 mmol/L β-mercaptoethanol), and the supernatants were subjected to assays for GUS enzyme activity using 2 mM 4-methylumbelliferyl glucuronide (MUG) as substrate. The intensity of the fluorescence was measured at OD455, using 4-methylumbelliferone (MU) as an internal control. Protein concentration was determined according to Bradford (1976).
Results
Prediction of Embryo-Specific Elements in the Emb5 Promoter
We first cloned the full-length proEmb5 from genomic DNA isolated from the maize inbred line Qi319. Sequencing results showed that the homology between our sequence and the reported sequence (Kriz et al. 2006, patent No. US 7,078,234 B2) was 99.52%, where eight different bases were found at positions 240, 257, 286, 287, 388, 978, 998, and 1617 as indicated in Suppl Fig. 1S (boxed). This discrepancy could be due to the different maize plant inbred lines used between these two studies.
The promoter sequence was analyzed by the PLACE (Higo et al. 1999) and PlantCARE (Lescto et al. 2002) software, and the result is shown in Suppl Table 2S. It was found that the CAAT box and TATA box were present at 226 bp and 119 bp upstream of the transcription start codon. Many cis-regulatory elements that are predicted to be embryo-specific expression elements were also found. These include the Skn-1-motif (− 1460 bp, − 1632 bp), numerous G-boxes, E-boxes (− 132 bp, − 508 bp, − 704 bp, − 859 bp, − 907 bp, − 1117 bp, and − 1623 bp), C-box (− 993 bp), pyrimidine box (− 730 bp), − 300 element (− 372 bp), TGTTG-motif (− 1347 bp), ATTTTC-motif (− 1432 bp), and TGAATG-motif (− 1545 bp). Interestingly, we also noted seven conserved elements of 5′-CG/T/C/T/A/CGTGT-3′ at positions of − 925 bp, − 899 bp, − 674 bp, − 634 bp, − 284 bp, − 120 bp, and − 65 bp (Suppl Table 2S; Suppl Fig. 2S.
The Emb5 promoter also contains several possible transcriptional enhancing elements, such as AT-rich motif (− 1611 bp, − 1577 bp, − 1556 bp, − 1516 bp, − 1470 bp, − 1419 bp, − 1358 bp, − 1210 bp, − 769 bp, − 400 bp, − 145 bp), AGCCCA-motif (− 1409 bp), GAAAAA-motif (− 1299 bp), CAAT-box (− 1645 bp, − 1535 bp, − 1510 bp, and − 1476 bp), CAAT-like box (− 430 bp), CACT-motif (− 1340 bp, − 1322 bp, − 1235 bp, − 469 bp), and GCAA-motif (− 1558 bp, − 1566 bp, − 1558 bp, − 1536 bp). In addition, many biotic stress and hormone response elements, such as the ABA response element ABRE (− 495 bp) and SA response element TCA (− 1365 bp) were also found.
Therefore, our data revealed that the Emb5 promoter contains a number of embryo-specific expression-related elements.
The Emb5 Promoter Had High Embryo-Specific Expression Activity in Arabidopsis
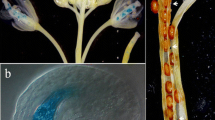
We first performed GUS histochemical staining assays on the transgenic Arabidopsis plants harboring the proEmb5:GUS (Emb5) and pro35S:GUS (35S) constructs. The results show that only the siliques of the proEmb5:GUS transgenic plants were stained blue, while other tissues such as the root, stem, leaves, and inflorescence were not stained (Fig. 1a; Fig.1b). Therefore, the full-length promoter could not initiate the expression of the GUS gene in tissues other than the silique, confirming the previous finding of the embryo-specific expression pattern of this promoter.
Detection of GUS expression and activity in different tissues of transgenic Arabidopsis plants. a GUS staining on roots, stems, leaves, inflorescences, and siliques of the transgenic plants expressing porEmb5:GUS. Siliques of the wild type (WT) and transgenics expressing the empty pBI121 vector (35S:GUS) were used as negative and positive controls, respectively. b GUS staining of seeds from WT, transgenic plants expressing the empty pBI121 vector (35S:GUS), and proEmb5:GUS, respectively. c Fluorescence measurements of GUS activity in WT, pBI121, and porEmb5:GUS transformed plants. Error bars represent means ± standard deviation of 3 replicates
The GUS activity in the transgenic plants was also quantified from four plants of each line after MUG staining. Figure 1 c shows that while GUS activity was hardly detected in tissues of the WT plants, it was detected in the transgenic plants of both 35S:GUS and Emb5:GUS. Interestingly, the highest expression levels were found in the seeds where the GUS activity of Emb5:GUS transgenic seeds exhibited 1.33 times higher than that of the 35S:GUS seeds.
The combined results clearly demonstrated that Emb5 could drive gene expression specifically in seeds of Arabidopsis.
Next, we determined the temporal expression activity of the Emb5 promoter during seed development by both histochemical staining and fluorescence quantification analysis of GUS activity in the seeds of Emb5:GUS transgenic plants at 5–9 days, 11 days, 12 days, and 14 days after flowering (DAF). Histochemical staining showed that GUS began to express in the seeds at around 9 DAF and the expression level reached the highest at 12 DAF (Fig. 2a). Soon after that, it started to decrease, and at 18 DAF, hardly any signal was detected in the transgenic seeds (Fig. 2b, c).
proEmb5 can drive embryo-specific expression of GUS. a Histochemical staining of the immature embryos during the late embryogenesis at 9, 11, 12, and 14 days after flowering (DAF) in the seeds of transgenic Arabidopsis plants expressing proEmb5:GUS. b Quantitative measurement of GUS activities during seed development. Error bars represents means ± standard deviation of 3 replicates. c Fold change of GUS activity in transgenic embryos at different stages of their development using 1 DAF as the reference
Analysis of the Core Functional Regions of Emb5 Promoter Associated with Its Embryo-Specific Activity
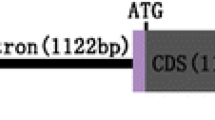
In order to determine the core functional region(s) of the embryo-specific activity, we subsequently constructed the series of 5′ deletion constructs in the plant expression vector pBI121, which has the GUS reporter gene sequence located downstream of the 35S constitutive promoter. By replacing the original 35S promotor with these fragments, we constructed proEmb5A (− 1143 bp ~ − 1 bp), proEmb5B (− 956 bp ~ − 1 bp), proEmb5C (− 732 bp ~ − 1 bp), proEmb5D (− 523 bp ~ − 1 bp), proEmb5E (− 331 bp ~ − 1 bp), proEmb5F (− 251 bp ~ − 1 bp), and proEmb5G (− 127 bp ~ − 1 bp), respectively (Fig. 3). Transgenic lines containing these constructs were generated, and GUS activity was subsequently monitored in the seeds of the transgenic Arabidopsis plants.
Schematic diagram of the Emb5 promoter structure and the seven 5′-flanking deletion fragments. The full lengths of proEmb5, proA, proB, proC, proD, proE, proF, and proG are 1.6, 1.14, 0.95, 0.73, 0.52, 0.33, 0.25, and 0.12 kb, respectively. The positions and numbers of the G-boxes (orange), CAAT-box (green), and E-box (pink) in each fragment are indicated
The histochemical staining assays showed that all the promoter fragments, except proEmb5-G, could drive GUS expression. Interestingly, the 523 bp proEmb5D between − 523 bp and ~ − 1 bp showed the darkest blue stain, which was comparable with the full-length proEmb5. However, the shortest fragment proEmb5-G between − 127 bp and ~ − 1 bp completely lost the promoter activity as the embryo did not stain at all. Quantitative data showed a similar trend where 85, 95, 100, 205, 80, 82, and 25 pmol/mμ/mg protein/min were detected in the seeds of proEmb5A-, B-, C-, D-, E-, F-, and G-GUS expression lines, respectively, where the highest value of 205 pmol/mμ/mg protein/min for proEmb5D expressing seeds was obtained (Fig. 4a–c). Therefore, the combined data indicate that the core functional element of proEmb5 lies between − 523 bp and ~ − 331 bp (between proEmb5D, − 523 bp ~ − 1 bp, and proEmb5E, − 331 bp ~ − 1 bp).
GUS activity of different deletion fragments of the proEmb5 in transgenic Arabidopsis plants. a GUS staining of transgenic embryos at the late embryogenesis. b GUS activities quantified from embryos in a. Error bars represents means ± standard deviation of 3 replicates. c Fold changes of GUS activity calculated from data in B using WT as the reference
Because very young seedlings after germination were the continuous tissues of embryos, the strong GUS stain should indicate embryo specificity of the full-length and shorter promoters; we further analyzed the 1–10 days old seedlings of the above transgenic lines for the temporal expression of GUS. As shown Fig. 5 that apart from the shortest fragment pG, which lost promoter activity altogether, all other shorter promoter fragments maintained the embryo specificity because only seeds and very young seedlings before 3 DAF were stained. Importantly, this followed the same trend as the full-length promoter pEm-GUS where its embryo specificity had already confirmed previously and in this study (Fig. 1). However, the GUS stain became lighter after 3 days when they were transited to vegetative tissues until disappeared completely from 8 day-old pA,B,E,F-GUS seedlings and 10 day-old pEm-, pC-, and pD-GUS seedlings, while seedlings from p35S-GUS were stained blue throughout this period and beyond.
GUS staining of transgenic Arabidopsis seedlings expressing the pEmb5:GUS and the seven 5′-deletion constructs. Seedlings at 1, 3, 5 (proE:GUS), 6, 8, and 10 days after germination were stained and observed. Transgenic seedlings expressing the empty pBI121 vector (p35S::GUS) were used as the positive control
Taken together, these results demonstrated that except for pG, which has very little if any promoter activity, the full length of Emb5 and all other remaining 5′ deletion fragments have embryonic specificity, although they have different transcriptional activities. Therefore, this further validated the results obtained from GUS assays performed on the immature embryos of these shown in Fig. 4a–c.
Identification of the Positive Regulatory Elements for Embryo-Specific Activity Within the En and Dn Regions
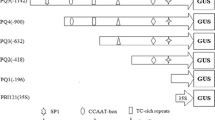
Bioinformatics analysis and GUS staining assays showed that the regions between − 1653 bp ~ − 1143 bp (510 bp, named hereafter En) and − 523 bp ~ − 331 bp (192 bp, named hereafter Dn) may contain the embryo-specific enhancer elements (Table 2S; Fig. 2S; Fig. 4a–c). In order to further verify this and to identify the possible positive regulatory elements within these two fragments, they were sub-cloned in the pMD-18-T-simple vector. The En fragment was again divided into two nearly equal sized fragments of En-1 (− 1653 bp ~ − 1385, 268 bp) and En-2 (− 1385 bp ~ − 1143 bp, 242 bp). All these four fragments were fused to the 35S minimal promoter and cloned in pBI121 to result in the construction of pBI121-En-35Smini:GUS, pBI121-Dn-35Smini:GUS, pBI121-En-1-35Smini:GUS, and pBI121-En-2-35Smini:GUS plant expression vectors, respectively (Fig. 6).
Maps of the seven GUS reporter expression vectors containing the two positive regulatory regions of the Emb5 promoter. The positions of the regions in relation to the full-length proEmb5 are indicated. En: − 1653 bp ~ − 1143 bp, Dn: − 523 bp ~ − 331 bp), NA (− 127 bp ~ − 1 bp, no GUS activity), 35S: CaMV 35S minimal promoter
Following the transformation of Arabidopsis, the developing seeds from all transgenic lines containing the above constructs were again assessed for GUS activity by histochemical staining as well as fluorescent quantification on the 12th day after flowering. As shown in Fig. 7a, the En-35Smp-GUS and Dn-35Smp-GUS transgenic seeds were stained light blue while the negative control (35Smini-GUS) was not stained at all. Measurements of the fluorescence activity showed that at 70 pmol/mμ/mg protein/min, the intensity of the fluorescence generated by En-35Smp-GUS seeds was 0.95 times higher than that of Dn-35Smp-GUS. Transgenic seeds containing the two smaller fragments of En, En-1-35Smp, and En-2-35Smini did not show much staining. Consistent with this, the fluorescent intensity was much lower than En-35Smp-GUS seeds, and there was no significant difference found between them (both at ~ 45 pmol/mμ/mg protein/min) (Fig. 7b).
GUS activities of transgenic seeds harboring the different constructs containing the positive regulatory elements. a GUS staining of the developing seeds of WT and transgenic plants at 12 days after flowering. b GUS activity measured by the fluorescence in seeds corresponding to plant lines in a Error bars represent represents means ± standard deviation of 3 replicates
To see if the promoter fragments pD, Dn, and En of Emb5 have additive effects, we constructed vectors of En-Dn-35Smini:GUS, Dn-Dn-35Smini:GUS, and En-pD:GUS by overlapping PCR where En + Dn, Dn + Dn and En + pD were fused together. The full-length proEmb5-GUS was used as the positive control, and the 35Smini as the negative control. For the En-pD:GUS vector, the WT was used as the negative control. The results showed that plants containing the En-pD stained the deepest in dark blue, and this was in line with the highest fluorescence quantitative activity measurement, amounting 231 pmol/mμ/mg protein/min (Fig. 7a, b). This was followed by the plants harboring the full-length Emb5 promoter and pD where the fluorescence quantitative activity was similar at about 203 pmol/mμ/mg protein/min, although the former appeared to be slightly darker in the GUS staining assay (Fig. 7a, b). At 105 pmol/mμ/mg protein/min, the Dn-Dn-35Smp and En-Dn-35Smp transgenic plants were only about half of those containing the full-length Emb5 and pD promoters, and it appeared no significant difference between them (Fig. 7a, b).
Therefore, our data clearly demonstrated that the activity of combined En and pD promoter regions of the Emb5 promoter presented the highest activity towards embryo-specific expression of GUS, which was followed by pD in Arabidopsis. The activities of Dn-Dn and En-Dn were significantly higher than Dn and En alone. This indicates that the regions within En and Dn contain the positive regulatory elements, which can act additively to enhance the embryo-specific transcriptional activity of the Emb5 promoter. Therefore, with much shorter at 523 bp for pD and 1033 bp for pEn-pD than the full-length Emb5 promoter (1653 bp), these two fragments could be the choice of embryo-specific promoters to use in research and application in plant science and biotechnology.
Discussion
In this study, we further characterized the tissue specificity and expression activity of the embryo-specific promoter of the maize embryo-specific Emb5 gene in Arabidopsis, leading to the identification of a much shorter region of 523 bp that contains all the required cis-elements to drive embryo-specific expression of this promoter. This maize-derived short yet strong promoter activity that is comparable with the full-length promoter (1653 bp) could be used for seed-specific expression of genes of interest in monocots as well as in dicots. In addition, due to its short length, this newly identified sequence could become the promotor of choice in in vitro manipulation for pathway engineering and signaling pathways where multiple transgenes are required.
To determine the embryo-specific activity of the Emb5 promoter, we utilized the model plant Arabidopsis as a host to express the full-length as well as its 5′ deleted shorter DNA fragments, which were individually fused with the GUS reporter gene. This strategy has been used previously by many other researchers to dissect the functions of unknown promoters from various plant species. For example, Chung et al. (2008) noted that the promoter of PfOle19 gene from the monocotyledonous plant Perilla contains 7 embryo-specific cis-regulatory elements, such as AACAA and five E-boxes (CANNTG). GUS expression driven by this promoter in Arabidopsis leads to embryo-specific staining, confirming its expression activity towards embryo. Zavallo et al. (2010) pointed out that the embryo specificity of the HaFAD2–1 promoter of dicotyledonous sunflower was determined by four embryo-specific elements in its sequence, including one Sh1-box (TGAATG) and three E-boxes (CANNTG). Therefore, the dicotyledonous plant Arabidopsis can be used as the host to express and analyze the core functional regions of a promoter and its cis-regulatory elements of genes originated from both monocots and dicots.
We first carried out bioinformatics analysis and found that the maize Emb5 promoter also contains the embryo-specific elements, such as one Sh1-box (at position − 1538 bp) and seven E-boxes (at positions of − 132 bp, − 508 bp, − 704 bp, − 859 bp, − 907 bp, − 1117 bp, and − 1623 bp) (Fig. 2S). This suggests that this promoter may also have the ability to drive gene expression in embryos. Indeed, our GUS assays showed that the Emb5 promoter had the highest activity in the Arabidopsis embryos compared with other tissues (Fig. 1). This demonstrated that the Emb5 promoter could drive embryo-specific expression in Arabidopsis. The expression activity of the GUS gene driven by the Emb5 promoter in Arabidopsis was the highest during the mid- to late developmental stages of the embryos (Fig. 2). This was consistent with the expression pattern of the Emb5 gene in developing maize seeds (Williams and Tsang 2005). Therefore, the embryo-specific expression pattern of Emb5 in Arabidopsis was similar to that in maize, a monocotyledonous plant. These combined results, again, provided clear evidence that the dicotyledonous model plant Arabidopsis could be employed to study the core functional regions and cis-regulatory elements related to embryo-specific expression activity of this monocotyledonous maize Emb5 promoter.
Having confirmed the embryo-specific expression pattern of the full-length Emb5 promoter in Arabidopsis, we next studied the specific regions and cis-elements that are responsible for this activity. The first experiment we carried out was to construct a series of 5′-deletion fragments with each fragment of ~ 500 bp in length (Fig. 3). This revealed that the proD between − 523 bp and − 1 bp had an activity comparable with that of the full-length Emb5 promoter of 1653 bp, while all the other fragments had lower activity than the full-length Emb5 promoter (Fig. 4). Deletion of the 192 bp between − 523 bp and − 331 bp (Dn) within proD significantly reduced the activity, suggesting that this region may contain the cis-acting elements with enhanced promoter activity. On the contrary, the activity of GUS driven by proA, proB, and proC fragment was significantly lower than that of proD, indicating that there may be negative regulatory elements present between − 1143 bp ~ − 523 bp and − 732 bp ~ − 523 bp. Furthermore, the fact that the promoter activity of the full-length Emb5 fragment was significantly higher than that of the pA fragment (− 1143 bp ~ − 1 bp) indicates that there could be positive regulatory elements between − 1653 bp and − 1143 bp that can enhance transcriptional activity of the promoter.
It is interesting to note that the region within proD contains the CAAT-like box (at − 430) and an AT-rich element (at − 400), which are known to have enhanced transcriptional activity (Zhang et al. 2002). As the proA, proB, and proC fragments all showed a significant decrease in promoter activity compared with the proD fragment, we reasoned that the formers may contain negative regulatory elements that inhibit promoter activity. Further analysis revealed that there are three reverse complement motifs of 5′-ACACNNG-3′ present within these regions. These are CGAGTGT (− 674 bp ~ − 667 bp), CGTGTGT (− 898 bp ~ − 891 bp), and CGTGTGT (− 924 bp ~ − 917 bp), respectively. Finkelstein and Lynch (2000) demonstrated that the 5′-ACACNNG-3′ element is a novel bZIP-like transcription factor core-binding sequence that promotes specific expression of the ABI5 gene in embryos, and the absence of this binding region inhibits transcriptional activity. Therefore, these three reverse complement sequences (5′-CNNGTGT-3′) within the Emb5 promoter region could potentially impose a strong inhibitory effect on Emb5 expression in the embryo of maize. This may explain why the GUS activity was reduced under the control of the proA-C promotor regions in Arabidopsis (Fig. 4).
Similarly, high GUS activity was found with plants expressing the proEn:GUS because this region (between − 1653 bp and − 1143 bp) contains the transcriptional enhancing elements, such as the AT-rich motifs, AGCCCA-motif, GAAAAA-motif, CAAT-box, CACT-motif, and GCAA-motif (Fig. 2S). Therefore, proEn functions in a similar fashion as proDn to positively drive the high embryo-specific expression in Arabidopsis.
To see if En and Dn have an additive effect, we fused Dn and En and Dn with Dn to produce proDn-En:GUS and pro2xDn:GUS and introduced them in Arabidopsis plants. As shown in Fig. 7a, the GUS activity was much enhanced in the transgenic Arabidopsis plants expressing these constructs compared with that in the control plants harboring only one of the fragments. Using a similar strategy, the minimal active promotor region of the rice endosperm-specific promotor of rsu3 was mapped within the 700 bp fragment upstream of its translation initiation codon (Rasmussen and Donaldson 2006). In a separate study, Petersen et al. (1996) found that while a 22 bp W1 element in the promoter of the rice chitinase gene can hardly activate its expression, the multiple tandem copies of it can drive tissue-specific expression in plants.
It was disappointing that none of the shorter individual or the synthetic promotor fragments exhibited the same or stronger embryo-specific activity than the full-length Emb5 promotor. The reason for this could be because the spacing between the binding site(s) of corresponding transcription factor(s) and these fragments was altered in the new constructs. This could have some negative effect on the binding to transcription factors, thus affecting the transcription efficiency of GUS. This is in agreement with the study of the 1.8-kb promotor fragment of nitrate reductase gene NIA1 where the authors found that the enhancer activity was localized to a 180-bp fragment. Three cis-elements corresponding to predicted binding motifs for homeodomain/E-box, Myb, and Alfin1 transcription factors were identified within this 180-bp fragment. However, in order to obtain a fully active synthetic promoter built from these three elements, the spacing between the homeodomain/E-box and Myb-Alfin1 sites has to be maintained to that of the native promoter (Rongchen Wang et al. 2010). Therefore, it seems that the spacing between the regulating elements in the promoters and the downstream genes is very important for the promoter activities (Blackwood and Kadonaga 1998).
In summary, through sequence analysis and expression studies of the GUS fusion constructs in Arabidopsis, we further verified the embryo-specific expression pattern of the maize Emb5 promotor. Importantly, we identified the regions that are responsible for this activity and confirmed that the 523-bp proD can replace the 1653-bp full-length promotor of Emb5 to drive the high-level and embryo-specific expression in Arabidopsis seed. Because it has comparable expression activity with the full-length promoter while it is much shorter, it could be utilized for genetic improvement of yield and quality traits in crop plants, such as cereal crops, as well as other pathway engineering studies in the future.
References
Battraw MJ, Hall TC (1990) Histochemical analysis of CaMV 35S promoter-beta-glucuronidase gene expression in transgenic rice plants. Plant Mol Biol 15:527–538
Blackwood EM, Kadonaga JT (1998) Going the distance: a current view of enhancer action. Science 281:61–63
Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cheon BY, Kim HJ, Oh KH, Bahn SC, Ahn JH, Choi JW, Ok SH, Bae JM, Shin JS (2004) Overexpression of human erythropoietin (EPO) affects plant morphologies: retarded vegetative growth in tobacco and male sterility in tobacco and Arabidopsis. Transgenic Res 13:541–549
Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18:675–689
Chung KJ, Hwang SK, Hahn BS, Kim KH, Kim JB, Kim YH, Yang JS, HaKim SH (2008) Authentic seed-specific activity of the Perilla oleosin 19 gene promoter in transgenic Arabidopsis. Plant Cell Rep 27:29–37
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Drakakaki G, Christou P, Stöger E (2000) Constitutive expression of soybean ferritin cDNA in transgenic wheat and rice results in increased iron levels in vegetative tissues but not in seeds. Transgenic Res 9:445–452
Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12:599–609
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database Nucleic Acids Res 27:297–300
Jefferson R (1987) GUS: histochemical staining with X-Gluc. Plant Mol Biol Report 5:387–405
Kriz AL, Ferry GCT, Voyles DA, Pawcatuck CT (2006) Maize embryo-specific promoter, composition and methods for use thereof. Patent No.: US 7,078,234 B2, Date of Patent: Jul.18, 2006
Li T, Sun J, Li JT (2015) Research progress on plant promoter. Biotechnol Bull 31:18–25
Liu X, Yang W, Li Y, Li S, Zhou X, Zhao Q, Fan Y (2016) The intergenic region of the maize defensin-like protein genes Def1 and Def2 functions as an embryo-specific asymmetric bidirectional promoter. J Exp Bot 67:4403–4413
Naoumkina M, Dixon RA (2011) Characterization of the mannan synthase promoter from guar (Cyamopsis tetragonoloba). Plant Cell Rep 30:997–1006
Naoumkina M, Vaghchhipawala S, Tang Y, Ben Y, Powell RJ, Dixon RA (2008) Metabolic and genetic perturbations accompany the modification of galactomannan in seeds of Medicago truncatula expressing mannan synthase from guar (Cyamopsis tetragonoloba L.). Plant Biotechnol J 6:619–631
Pan Y, Ma X, Liang H, Zhao Q, Zhu D, Yu J (2015) Spatial and temporal activity of the foxtail millet (Setaria italica) seed-specific promoter pF128. Planta 241:57–67
Rasmussen TB, Donaldson IA (2006) Investigation of the endosperm-specific sucrose synthase promoter from rice using transient expression of reporter genes in guar seed tissue. Plant Cell Rep 25:1035–1042
Requesens DV, Romero MEG, Devaiah SP, Chang YK, Flory A, Streatfield S, Ring R, Phillips C, Hood NC, Marbaniang CD, Howard JA, Hood EE (2019) The maize α-zein promoter can be utilized as a strong inducer of cellulase enzyme expression in maize kernels. Transgenic Res 28:537–547
Rongchen Wang R, Guan P, Chen M, Xing X, Zhang Y, Crawford NM (2010) Multiple regulatory elements in the Arabidopsis NIA1 promoter act synergistically to form a nitrate enhancer. Plant Physiol 154:423–432
Saidi Y, Schaefer DG, Goloubinoff P, Zryd JP, Finka A (2009) The CaMV 35S promoter has a weak expression activity in dark grown tissues of moss Physcomitrella patens. Plant Signal Behav 4:457–459
Sohrabi M, Zebarjadi A, Najaphy A, Kahrizi D (2015) Isolation and sequence analysis of napin seed specific promoter from Iranian rapeseed (Brassica napus L.). Gene 563:160–164
Sunkara S, Bhatnagarmathur P, Sharma KK (2014) Isolation and functional characterization of a novel seed specific promoter region from peanut. Appl Biochem Biotechnol 172:325–339
Williams BA, Tsang A (2005) Analysis of multiple classes of abscisic acid-responsive genes during embryogenesis in Zea mays. Genesis 15:415–424
Yamori W (2013) Improving photosynthesis to increase food and fuel production by biotechnological strategies in crops. Plant Biochem Physiol 1:1–3
Zavallo D, Bilbao ML, Hopp HE, Heinz R (2010) Isolation and functional characterization of two novel seed-specific promoters from sunflower (Helianthus annuus L.). Plant Cell Rep 29:239–248
Zhang J, Li L, Song Y (2002) Identification of seed-specific promoter nap300 and its comparison with 7S promoter. Prog Nat Sci 12:737–741
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key message
A 523 bp DNA fragment was identified from Zea mays that can drive strong embryo-specific expression in Arabidopsis. Due to its small size, this promoter provides great versatility in vector construction to assemble multiple transgenes for efficient expression in seeds, for example.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Y., Yu, L., Wang, Q. et al. Analysis of the Promoter of Emb5 from Zea mays Identifies a Region of 523 bp Responsible for Its Embryo-Specific Activity. Plant Mol Biol Rep 39, 288–300 (2021). https://doi.org/10.1007/s11105-020-01251-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01251-w