Abstract
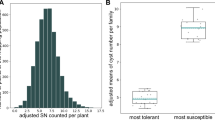
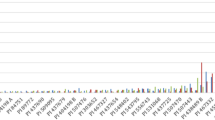
The beet-cyst nematode (Heterodera schachtii Schmidt) is one of the major pests of sugar beet. The identification of molecular markers associated with nematode tolerance would be helpful for developing tolerant varieties. The aim of this study was to identify single nucleotide polymorphism (SNP) markers linked to nematode tolerance from the Beta vulgaris ssp. maritima source WB242. A WB242-derived F2 population was phenotyped for host-plant nematode reaction revealing a 3:1 segregation ratio of the tolerant and susceptible phenotypes and suggesting the action of a gene designated as HsBvm-1. Bulked segregant analysis (BSA) was used. The most tolerant and susceptible individuals were pooled and subjected to restriction site associated DNA sequencing (RAD-Seq) analysis, which identified 7,241 SNPs. A subset of 384 candidate SNPs segregating between bulks were genotyped on the 20 most-tolerant and most-susceptible individuals, identifying a single marker (SNP192) showing complete association with nematode tolerance. Segregation of SNP192 confirmed the inheritance of tolerance by a single gene. This association was further validated on a set of 26 commercial tolerant and susceptible varieties, showing the presence of the SNP192 WB242-type allele only in the tolerant varieties. We identified and mapped on chromosome 5 the first nematode tolerance gene (HsBvm-1) from Beta vulgaris ssp. maritima and released information on SNP192, a linked marker valuable for high-throughput, marker-assisted breeding of nematode tolerance in sugar beet.


Similar content being viewed by others
Abbreviations
- SNP:
-
Single nucleotide polymorphisms
- RAD-Seq:
-
Restriction site associated DNA sequencing
- BSA:
-
Bulk segregant analysis
References
Barchi L, Lanteri S, Portis E, Acquadro A, Valè G, Toppino L, Rotino GL (2011) Identification of SNP and SSR markers in eggplant using RAD tag sequencing. BMC Genomics 12:304
Biancardi E, Lewellen RT, De Biaggi M, Erichsen AW, Stevanato P (2002) The origin of rhizomania resistance in sugar beet. Euphytica 127:383–397
Biancardi E, McGrath JM, Panella LW, Lewellen RT, Stevanato P (2010) Sugar beet. In: Bradshaw J (ed) Handbook of plant breeding, vol 4, Tuber and Root Crops. Springer, New York, pp 173–219
Biancardi E, Panella LW, Lewellen RT (2012) Beta maritima: the origin of beets Springer-Verlag New York Inc., pp 293
Brandes A, Jung C, Wricke G (1987) Nematode resistance derived from wild beet and its meiotic stability in sugar beet. Plant Breed 99:56–64
Cai D, Kleine M, Kifle S, Harloff HJ, Sandal NN, Marcker KA, Klein-Lankhorst RM, Salentijn EM, Lange W, Stiekema WJ, Wyss U, Grundler FM, Jung C (1997) Positional cloning of a gene for nematode resistance in sugar beet. Science 275:832–834
Chutimanitsakun Y, Nipper RW, Cuesta-Marcos A, Cistué L, Corey A, Filichkina T, Johnson EA, Hayes PM (2011) Construction and application for QTL analysis of a restriction site associated DNA (RAD) linkage map in barley. BMC Genomics 12:4
Conover WJ (1980) Practical non-parametric statistics. Wiley, New York, p 592
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12:499–510
Jung C, Cai D, Kleine M (1998) Engineering nematode resistance in crop species. Trends Plant Sci 3:266–271
Kleine M, Voss H, Cai D, Jung C (1998) Evaluation of nematode-resistant sugar beet (Beta vulgaris L.) lines by molecular analysis. Theor Appl Gen 97:896–904
McCarter JP (2008) Molecular approaches toward resistance to plant-parasitic nematodes. In: Plant cell monographs. Springer, Berlin, pp 1–29
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA (2007) Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res 17:240–248
Panella L, Lewellen RT (2007) Broadening the genetic base of sugar beet: introgression from wild relatives. Euphytica 154:383–400
Pegadaraju V, Nipper R, Hulke B, Qi L, Schultz Q (2013) De novo sequencing of sunflower genome for SNP discovery using RAD (Restriction site Associated DNA) approach. BMC Genomics 14:556
Pelsy F, Merdinoglu D (1996) Identification and mapping of random amplified polymorphic DNA markers linked to a rhizomania resistance gene in sugar beet (Beta vulgaris L.) by bulked segregant analysis. Plant Breed 115:371–377
Stevanato P, Trebbi D, Saccomani M (2010) Root traits and yield in sugar beet: identification of AFLP markers associated with root elongation rate. Euphytica 173:289–298
Stevanato P, Broccanello C, Biscarini F, Del Corvo M, Panella L, Gaurav S, Stella A, Concheri G (2013) First application of QuantStudio platform for high throughput SNP marker assessment in sugar beet. Plant Mol Biol Rep. doi:10.1007/s11105-013-0685-x
Taguchi K, Okazaki K, Takahashi H, Kubo T, Mikami T (2010) Molecular mapping of a gene conferring resistance to Aphanomyces root rot (black root) in sugar beet (Beta vulgaris L.). Euphytica 173:409–418
Thurau T, Ye W, Menkhaus J, Knecht K, Tang G, Cai D (2010) Plant nematode control. Sugar Tech 12:229–237
Touzet P, Hueber N, Burkholz A, Barnes S, Cuguen J (2004) Genetic analysis of male fertility restoration in wild cytoplasmic male sterility G of beet. Theor Appl Genet 109:240–247
Van Ooijen JW (2011) Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Gen Res 93:343–349
Acknowledgments
The authors thanks Robert Lewellen and Enrico Biancardi for their insightful comments on this study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material S1
(DOC 29 kb)
Supplementary material S2
(DOC 27 kb)
Rights and permissions
About this article
Cite this article
Stevanato, P., Trebbi, D., Panella, L. et al. Identification and Validation of a SNP Marker Linked to the Gene HsBvm-1 for Nematode Resistance in Sugar Beet. Plant Mol Biol Rep 33, 474–479 (2015). https://doi.org/10.1007/s11105-014-0763-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0763-8