Abstract
Background and aims
Some elephant grass (Pennisetum purpureum) genotypes are able to produce large amounts of biomass and accumulate N derived from BNF when growing in soil with low N levels. However, information about the diazotrophic bacteria colonizing this C4 plant is still very scarce. This study aimed to characterize the plant growth promoting traits of a fraction of culturable diazotrophs colonizing the genotypes CNPGL F06-3 and Cameroon.
Methods
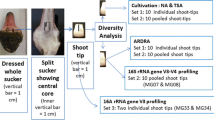
A total of 204 isolates were obtained from surface sterilized leaves, stems and roots after culturing on five different N-free semisolid media. These were then analyzed by BOX-PCR, and the 16S rRNA and nifH sequences of representative isolates were obtained. The functional ability of the isolates to reduce acetylene, produce indole and to solubilize phosphate was also determined.
Results
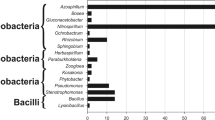
The diazotrophic bacterial population varied from 102 up to 106 bacteria g−1 fresh tissues of both genotypes. The BOX-PCR analysis suggested a trend in the genetic diversity among the 204 diazotrophic strains colonizing the different genotypes and plant tissues. Sequencing of 16S rRNA fragments confirmed the presence of Azospirillum brasilense and Gluconacetobacter diazotrophicus and revealed for the first time the occurrence of G. liquefaciens, G. sacchari, Burkholderia silvatlantica, Klebsiella sp., Enterobacter cloacae and E. oryzae in elephant grass. Interestingly, several nifH sequences from isolates identified as G. liquefaciens and G. sacchari showed homologies with nifH sequences of Enterobacter species. The majority of the isolates (97%) produced indole compounds, 22% solubilized phosphate and 6.4% possessed both characteristics.
Conclusions
The results showed the occurrence of novel diazotrophic bacterial species colonizing different tissues of both genotypes of elephant grass. In addition, the study revealed the presence of several bacteria with growth promoting traits, and highlighted their potential to be exploited as biofertilizers.


Similar content being viewed by others
References
Ahemad F, Ahmad I, Khan MS (2008) Screening of free-living rhizobacteria for their multiple plant growth promoting activities. Microbiol Res 163:173–181
Albino U, Saridakis DP, Ferreira MC, Hungria M, Vinuesa P, Andrade G (2006) High diversity of diazotrophic bacteria associated with the carnivorous plant Drosera villosa var. villosa growing in oligotrophic habitats in Brazil. Plant Soil 287:199–207
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Andrade AC, Fonseca DM, Lopes RS, Nascimento Júnior D, Cecon PR, Queiroz DS, Pereira DH, Reis ST (2005) Análise de crescimento do capim-elefante ‘napier’ adubado e irrigado. Ciênc agrotec 29:415–423
Baldani JI, Baldani VLD (2005) History on the biological nitrogen fixation research in graminaceous plants: special emphasis on the Brazilian experience. An Acad Bras Cienc 77:549–579
Baldani JI, Baldani VLD, Dobereiner J (2005) Genus III. Herbaspirillum. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds) Bergey’s manual of systematic bacteriology, 2nd edn. Springer, Newark, pp 629–636
Bashan Y, Bashan LE (2010) How the plant growth-promoting bacterium Azospirillum promotes plant growth-a critical assessment. Adv Agron 108:77–136
Boddey RM (1987) Methods for quantification of nitrogen fixation associated with gramineae. Crit Rev Plant Sci 6:209–266
Boddey RM (1995) Biological nitrogen fixation in sugar cane: a key to energetically viable biofuel production. Crit Rev Plant Sci 14:263–279
Couillerot O, Poirier MA, Prigent-Combaret C, Mavingui P, Caballero-Mellado J, Moënne-Loccoz Y (2010) Assessment of SCAR markers to design real-time PCR primers for rhizosphere quantification of Azospirillum brasilense phytostimulatory inoculants of maize. J Appl Microbiol 109:528–538
Crozier A, Kamiya Y, Bishop G, Yokota T (2000) Biosynthesis of hormones and elicitor molecules. In: Buchanan B, Gruissem W, Jones R (Ed.) Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists pp 850–929.
Dastager SG, Deepa CK, Puneet SC, Nautiyal CS, Pandey A (2009) Isolation and characterization of plant growth-promoting strain Pantoea NII-186. From Western Ghat forest soil, India. Lett Appl Microbiol 49:20–25
Dobbelaere S, Vanderleyden J, Okon Y (2003) Plant growth promoting effects of diazotrophs in the rhizosphere. Crit Rev Plant Sci 22:107–149
Döbereiner J (1988) Isolation and identification of root associated diazotrophs. Plant Soil 110:207–212
Döbereiner J (1995) Isolation and identification of aerobic nitrogen-fixing bacteria from soil and plants. In: Alef K, Nannipieri P (eds) Methods in applied soil microbiology and biochemistry. Academic, London, pp 134–141
Eckert B, Weber OB, Kirchhof G, Halbritter A, Stoffels M, Hartmann A (2001) Azospirillum doebereinerae sp. nov., a nitrogen-fixing bacterium associated with the C4-grass Miscanthus. Int J Syst Evol Micr 51:17–26
Fischer D, Pfitzner B, Schmid M, Simões-Araújo JL, Reis VM, Pereira W, Ormeño-Orrillo E, Hofmann A, Martinez-Romero E, Baldani JI, Hartmann A (2011) Molecular characterisation of the diazotrophic bacterial community in uninoculated and inoculated field-grown sugarcane (Saccharum sp.). Plant Soil doi:10.1007/s11104-011-0812-0
Franke-Whittle IH, Fegan M, Hayward C, Leonard G, Stackebrandt E, Sly LI (1999) Description of Gluconacetobacter sacchari sp. nov., a new species of acetic acid bacterium isolated from the leaf sheath of sugar cane and from the pink sugar-cane mealy bug. Int J Syst Bacteriol 49:1681–1693
Franke-Whittle IH, O’Shea MG, Leonard GJ, Webb RI, Sly LI (2005) Investigation into the ability of Gluconacetobacter sacchari to live as an endophyte in sugarcane. Plant Soil 271:285–295
Furushita M, Shiba T, Maeda T, Yahata M, Kaneoka A, Takahashi Y, Trii K, Hasegawa T, Ohta M (2003) Similarity of tetracycline resistance genes applied isolated from fish farm bacteria to those from clinical isolates. Appl Environ Microb 69:5336–5342
Gyaneshwar P, James EK, Mathan N, Reddy PM, Reinhold-Hurek B, Ladha JK (2001) Endophytic colonization of rice by a diazotrophic strain of Serratia marcescens. J Bacteriol 183:2634–2645
Hartmann A, Bashan Y (2009) Ecology and application of Azospirillum and other plant growth-promoting bacteria (PGPB). Eur J Soil Biol 45:1–122
Hurek T, Wagner B, Reinhold-Hurek B (1997) Identification of N2-fixing plant- and fungus-associated Azoarcus species by PCR-based genomic fingerprints. Appl Environ Microb 63:4331–4339
Islam MR, Madhaiyan M, Boruah HPD, Yim W, Lee G, Saravanan VS, Fu Q, Hu H, Sa T (2009) Characterization of plant growth-promoting traits of free-living diazotrophic bacteria and their inoculation effects on growth and nitrogen uptake of crop plants. J Microbiol Biotechn 19:1213–1222
Jha B, Thakura MC, Gontia I, Albrecht V, Stoffels M, Schmid M, Hartmann A (2009) Isolation, partial identification and application of diazotrophic rhizobacteria from traditional Indian rice cultivars. Eur J Soil Biol 45:62–72
Juraeva D, George E, Davranov K, Ruppel S (2006) Detection and quantification of the nifH gene in shoot and root of cucumber plants. Can J Microbiol 52:731–739
Kaschuk G, Hungria M, Andrade DS, Campo RJ (2006) Genetic diversity of rhizobia associated with common bean (Phaseolus vulgaris L.) grown under no-tillage and conventional systems in Southern Brazil. Appl Soil Ecol 32:210–220
Khan MS, Zaidi A, Wani PA, Ahemad M, Oves M (2009) Functional diversity among plant growth-promoting rhizobacteria. In: Khan MS, Zaidi A, Musarrat J (eds) Microbial strategies for crop improvement. Springer, Berlin, pp 105–132
Kirchhof G, Reis VM, Baldani JI, Eckert B, Döbereiner J, Hartmann A (1997a) Occurrence, physiological and molecular analysis of endophytic diazotrophic bacteria in gramineous energy plants. Plant Soil 104:45–55
Kirchhof G, Schloter M, Abmus B, Hartmann A (1997b) Molecular microbial ecology approaches applied to diazotrophs associated with non-legumes. Soil Biol Biochem 29:853–862
Kirchhof G, Eckert B, Stoffels M, Baldani JI, Reis VM, Hartmann A (2001) Herbaspirillum frisingense sp. nov., a new nitrogen-fixing bacterial species that occurs in C4-fiber plants. Int J Syst Evol Microbiol 51:157–168
Lerner A, Valverde A, Castro-Sowinski S, Lerner H, Okon Y, Burdman S (2010) Phenotypic variation in Azospirillum brasilense exposed to starvation. Environ Microbiol Rep 2:1758–2229
Mehta S, Nautiyal CS (2001) An efficient method for qualitative screening of phosphate-solubilizing bacteria. Curr Microbiol 43:51–56
Moore FP, Barac T, Borremans B, Oeyen L, Vangronsveld J, van der Lelie D, Campbell D, Moore ERB (2006) Endophytic bacterial diversity in poplar trees growing on a BTEX-contaminated site: the characterisation of isolates with potential to enhance phytoremediation. Syst Appl Microbiol 29:539–556
Morais RF, Souza BJ, Leite JM, Soares LH, Alves BJR, Boddey RM, Urquiaga S (2009) Elephant grass genotypes for bioenergy production by direct biomass combustion. Pesq Agropec Bras 44:133–140
Morais RF, Quesada DM, Reis VM, Urquiaga S, Alves BJR, Boddey RM (2011) Contribution of biological nitrogen fixation to Elephant grass (Pennisetum purpureum Schum.). Plant Soil. doi:10.1007/s11104-011-0944-2
Nayak BS, Badgley B, Harwood VJ (2011) Comparison of genotypic and phylogenetic relationships of environmental Enterococcus isolates by BOX-PCR typing and 16S rRNA gene sequencing. Appl Environ Microbiol 77:5050–5055
Oliveira ALM, Urquiaga S, Döbereiner J, Baldani JI (2002) The effect of inoculating endophytic N2-fixing bacteria on micropropagated sugarcane plants. Plant Soil 242:205–215
Pariona-Llanos R, Ibañez De Santi Ferrara Felipe F, Soto Gonzales HH, Barbosa HR (2010) Influence of organic fertilization on the number of cultivable diazotrophic endophytic bacteria isolated from sugarcane. Eur J Soil Biol 46:387–393
Pedraza RO, RamíRez-Mata A, Xiqui M, Baca BE (2004) Aromatic amino acid aminotransferase activity and indole-3-acetic acid production by associative nitrogen-fixing bacteria. FEMS Microbiol Lett 233:15–21
Peng G, Zhang W, Luo H, Xie H, Lai W, Tan Z (2009) Enterobacter oryzae sp. nov., a nitrogen-fixing bacterium isolated from the wild rice species Oryza latifolia. Int J Syst Evol Microbiol 59:1650–1655
Perin L, Martinez-Aguilar L, Paredes Valdez G, Baldani JI, Estrada-De Los Santos P, Reis VM, Caballero-Mellado J (2006) Burkholderia silvatlantica sp. nov., a diazotrophic bacterium associated with sugarcane and maize. Int J Syst Evol Microbiol 56:1931–1937
Poly F, Monrozier LJ, Bally R (2001) Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152:95–103
Radwan TEE, Mohamed ZK, Reis VM (2002) Production of indole-3-acetic acid by different strains of Azospirillum and Herbaspirillum spp. Symbiosis 32:39–54
Raymond J, Siefert JL, Staples CR, Blankenship RE (2004) The natural history of nitrogen fixation. Mol Biol Evol 21:541–554
Reis VM, dos Reis FB Jr, Sales JF, Schloter M (2000) Characterisation of different polyclonal antisera to quantify Herbaspirillum spp. in Elephant grass (Pennisetum purpureum Schum). Symbiosis 29:139–150
Reis VM, Reis Junior FB, Quesada DM, Oliveira OCA, Alves BJR, Urquiaga S, Boddey RM (2001) Biological nitrogen fixation associated with tropical pasture grasses. Aust J Plant Physiol 28:837–844
Reis VM, Estrada-De Los Santos P, Tenorio-Salgado S, Vogel J, Stoffels M, Guyon S, Mavingui P, Baldani VLD, Schmid M, Baldani JI, Balandreau J, Hartmann A, Caballero-Mellado J (2004) Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int J Syst Evol Microbiol 54:2155–2162
Rodrigues E, Rodrigues LS, Oliveira ALM, Baldani VLD, Teixeira KRS, Urquiaga S, Reis VM (2008) Azospirillum amazonense inoculation: effects on growth, yield and N2 fixation of rice (Oryza sativa L.). Plant Soil 302:249–261
Rodríguez H, Fraga R (1999) Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv 17:319–339
Roesch LFW, Olivares FL, Passaglia LMP, Selbach PA, Sa ELS, Camargo FAO (2006) Characterization of diazotrophic bacteria associated with maize: effect of plant genotype, ontogeny and nitrogen supply. World J Microb Biot 22:967–974
Roy BD, Deb B, Sharma GD (2010) Role of acetic acid bacteria in biological N2 fixation- a review. Biofrontiers 1:47–57
Samson R, Mani S, Boddey R, Sokhansanj S, Quesada D, Urquiaga S, Reis V, Ho Lem C (2005) The potential of C4perennial grasses for developing a global bioheat industry. Crit Rev Plant Sci 24:461–495
Sarwar M, Kremer RJ (1995) Enhanced suppression of plant growth through production of L-tryptophan-derived compounds by deleterious rhizobacteria. Plant Soil 172:261–269
Sashidhar B, Podile AR (2010) Mineral phosphate solubilization by rhizosphere bacteria and scope for manipulation of the direct oxidation pathway glucose dehydrogenase. J Appl Microbiol 109:1–12
Spaepen S, Vanderleyden J, Okon Y (2009) Plant growth-promoting actions of rhizobacteria. Adv Bot Res 7:283–320
Taghavi S, Garafola C, Monchy S, Newman L, Hoffman A, Weyens N, Barac T, Vangronsveld J, Van Der Lelie D (2009) Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl Environ Microb 75:748–757
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Vassilev N, Vassileva M (2003) Biotechnological solubilization of rock phosphate on media containing agro-industrial wastes. Appl Microbiol Biot 61:435–440
Versalovic J, Schneider M, De Bruijn FJ, Lupski JR (1994) Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Meth Mol Cell Biol 5:25–40
Videira SS, Araújo JLS, Rodrigues LS, Baldani VLD, Baldani JI (2009) Occurrence and diversity of nitrogen-fixing Sphingomonas bacteria associated with rice plants grown in Brazil. FEMS Microbiol Lett 293:11–19
Wang RF, Cao WW, Cerniglia CE (1996) Phylogenetic analysis of Fusobacterium prausnitzii based upon the 16S rRNA gene sequence and PCR confirmation. Int J Syst Bacteriol 46:341–343
Xin G, Zhang GY, Kang JW, Staley JT, Doty SL (2009) A diazotrophic, indole-3-acetic acid-producing endophyte from wild cottonwood. Biol Fertil Soils 45:669–674
Yamada Y, Hoshino K, Ishikawa T (1997) The phylogeny of acetic acid bacteria based on the partial sequences of 16 s ribosomal RNA: the elevation of the subgenus Gluconoacetobacterium to generic level. Biosci Biotech Bioch 61:1244–1251
Acknowledgment
The authors thank Embrapa Agrobiologia, the project CNPq/INCT-FBN, Faperj, CNPq (edital Universal) for financial support and CAPES and CNPq for the fellowship of the first and last two authors. The authors also thank the colleagues Robert M. Boddey and David Johnston-Monje for proof reading this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Euan K. James.
Electronic supplementary material
Below is the link to the electronic supplementary material.

Figure S1
Dendrogram obtained by cluster analysis of the BOX-PCR data for isolates from JNFb and NFb media. Similarities were calculated using the Dice coefficient and the data clustered using the unweighted pair group method of averages (UPGMA). Type strains are included (✰). Isolates indicated with the symbol “*” were used for partial sequencing of 16S rRNA while those indicated with “★” were used for total sequencing of 16S rRNA. (JPEG 137 kb)

Figure S2
Dendrogram obtained by cluster analysis of the BOX-PCR data for isolates from LGI medium. Similarities were calculated using the Dice coefficient and the data clustered using the unweighted pair group method of averages (UPGMA). Type strains are included (✰). Isolates indicated with the symbol “*” were used for partial sequencing of 16S rRNA while those indicated with “★” were used for total sequencing of 16S rRNA. (JPEG 69 kb)

Figure S3
Dendrogram obtained by cluster analysis of the BOX-PCR data for isolates from LGI-P medium. Similarities were calculated using the Dice coefficient and the data clustered using the unweighted pair group method of averages (UPGMA). Type strains are included (✰). Isolates indicated with the symbol “*” were used for partial sequencing of 16S rRNA while those indicated with “★” were used for total sequencing of 16S rRNA. (JPEG 106 kb)

Figure S4
Dendrogram obtained by cluster analysis of the BOX-PCR data for isolates from JMV medium. Similarities were calculated using the Dice coefficient and the data clustered using the unweighted pair group method of averages (UPGMA). Type strains are included (✰). Isolates indicated with the symbol “*” were used for partial sequencing of 16S rRNA while those indicated with “★” were used for total sequencing of 16S rRNA. (JPEG 146 kb)
Rights and permissions
About this article
Cite this article
Videira, S.S., de Oliveira, D.M., de Morais, R.F. et al. Genetic diversity and plant growth promoting traits of diazotrophic bacteria isolated from two Pennisetum purpureum Schum. genotypes grown in the field. Plant Soil 356, 51–66 (2012). https://doi.org/10.1007/s11104-011-1082-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-011-1082-6