Abstract
Key message
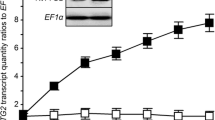
NtARF6 overexpression represses nicotine biosynthesis in tobacco. Transcriptome analysis suggests that NtARF6 acts as a regulatory hub that connect different phytohormone signaling pathways to antagonize the jasmonic acid-induced nicotine biosynthesis.
Abstract
Plant specialized metabolic pathways are regulated by a plethora of molecular regulators that form complex networks. In Nicotiana tabacum, nicotine biosynthesis is regulated by transcriptional activators, such as NtMYC2 and the NIC2-locus ERFs. However, the underlying molecular mechanism of the regulatory feedback is largely unknown. Previous research has shown that NbARF1, a nicotine synthesis repressor, reduces nicotine accumulation in N. benthamiana. In this study, we demonstrated that overexpression of NtARF6, an ortholog of NbARF1, was able to reduce pyridine alkaloid accumulation in tobacco. We found that NtARF6 could not directly repress the transcriptional activities of the key nicotine pathway structural gene promoters. Transcriptomic analysis suggested that this NtARF6-induced deactivation of alkaloid biosynthesis might be achieved by the antagonistic effect between jasmonic acid (JA) and other plant hormone signaling pathways, such as ethylene (ETH), salicylic acid (SA), abscisic acid (ABA). The repression of JA biosynthesis is accompanied by the induction of ETH, ABA, and SA signaling and pathogenic infection defensive responses, resulting in counteracting JA-induced metabolic reprogramming and decreasing the expression of nicotine biosynthetic genes in vivo. This study provides transcriptomic evidence for the regulatory mechanism of the NtARF6-mediated repression of alkaloid biosynthesis and indicates that this ARF transcription factor might act as a regulatory hub to connect different hormone signaling pathways in tobacco.







Similar content being viewed by others
Data availability
Sequencing data for root tissue RNA from the EV control line (SAMN18145490), three NtARF6-OE lines (SAMN18145491, SAMN18145492, and SAMN18145493 of N. tabacum cv. Coker176 were available from BioProject PRJNA706840 (https://www.ncbi.nlm.nih.gov/bioproject/). The genome and gene model of N. tabacum cv. K326 used for RNA-sequencing analysis were available at ftp://ftp.solgenomics . net/genomes/Nicotiana_tabacum/edwards_et_al_2017/assembly/Ntab-v4.5_genome_Scf_Edards2017.fasta.gz and ftp://ftp.solgenomics.net/genomes/Nicotiana_tabacum/edwards_et_al_2017/annotation/Ntab-v4.5_gene_models_Scf_Edwards2017.gff , respectively. The differential expressed genes and enriched KEGG pathways are included in this published article and its supplementary information files.
References
Anders A, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 18 July 2021
Baldwin IT, Schmelz EA, Ohnmeiss TE (1994) Wound-induced changes in root and shoot jasmonic acid pools correlate with induced nicotine synthesis in Nicotiana sylvestris spegazzini and comes. J Chem Ecol 20:2139–2157
Chen Q, Sun JQ, Zhai QZ, Zhou WK, Qi LL, Xu L, Wang B, Chen R, Jiang HL, Qi J, Li XG, Palme K, Li CY (2011) The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23:3335–3352
Chini A, Gimenez-Ibanez S, Goossens A, Solano R (2016) Redundancy and specificity in jasmonate signaling. Curr Opin Plant Biol 33:147–156
Chow C, Lee T, Hung Y, Li G, Tseng K, Liu Y, Kuo P, Zheng H, Chang W (2019) PlantPan3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acid Res 47:1155–1163
Clarke JD, Liu Y, Klessig DF, Dong X (1998) Uncoupling PR gene expression from NPR1 and bacterial resistance: characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10:557–569
Colinas M, Goossens A (2018) Combinatorial transcriptional control of plant specialized metabolism. Trends Plant Sci 23:324–336
Edwards KD, Fernandez-Pozo N, Drake-Stowe K, Humphry M, Evans AD, Bombarely A, Allen F, Hurst R, White B, Kernodle SP, Bromley JR, Sanchez-Tamburrino JP, Lewis RS, Mueller LA (2017) A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genom 18:448
Guifoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10:453–460
Gupta V, Willits MG, Glazebrook J (2000) Arabidopsis thaliana EDS4emclose contributes to salicylic acid (SA)-dependent expression of defense responses: evidence for inhibition of jasmonic acid signaling by SA. Mol Plant-Microbe Interact 13:503–511
Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF (2001) A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc Natl Acad Sci USA 98:9448–9453
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN (2001) Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J 26:509–522
Lackman P, González-Guzmán M, Tilleman S, Carqueijeiro I, Pérez AC, Moses T, Seo M, Kanno Y, Hakkinen S, Van Montagu MCE, Thevelein JM, Maaheimo H, Oksman-Caldentey K, Rodriguez PL, Rischer H, Goossens A (2011) Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc Natl Acad Sci USA 108:5891–5896
Laudert D, Weiler EW (1998) Allene oxide synthase: a major control point in Arabidopsis thaliana octadecanoid signaling. Plant J 15:675–684
Leon-Reyes A, Van der Does D, De Lange ES, Delker C, Wasternack C, Van Wees SCM, Ritsema T, Pieterse CMJ (2010) Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 232:1423–1432
Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16:319–331
Li K, Wang S, Wu H, Wang H (2020) Protein levels of several Arabidopsis auxin response factors are regulated by multiple factors and ABA promotes ARF6 protein ubiquitination. Int J Mol Sci 21:9437
Liu N, Wu S, Van Houten J, Wang Y, Ding B, Fei Z, Clarke TH, Reed J, van der Knaap E (2014) Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to flora development defects and female sterility in tomato. J Exp Bot 65:2507–2520
Lu X, Zhang L, Zhang F, Jiang W, Shen Q, Zhang L, Lv Z, Wang G, Tang K (2013) AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrtis cinereal. New Phytol 198:1191–1202
Roosjen M, Paque S, Weijers D (2018) Auxin response factors: output control in auxin biology. J Exp Bot 69:178–188
Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, Ecker JR, Reed JW (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132:4107–4118
Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C (2007) SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF12 transcription. Plant J 50:128–139
Nugroho LH, Peltenburg-Looman AMG, de Vos H, Verberne MC, Verpoorte R (2002) Nicotine and related alkaloids accumulation in constitutive salicylic acid producing tobacco plants. Plant Sci 162:575–581
Paul P, Singh SK, Patra B, Sui X, Pattanaik S, Yuan L (2017) A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol 213:1107–1123
Paul P, Singh SK, Patra B, Liu X, Pattanaik S, Yuan L (2020) Mutually regulated AP2/ERF gene clusters modulate biosynthesis of specialized metabolites in plants. Plant Physiol 182:840–856
Pauw B, Hilliou FAO, Martin VS, Chatel G, de Wolf CJF, Champion A, Pré M, van Duijn B, Kijne JW, van der Fits L, Memelink J (2004) Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. J Biol Chem 279:52940–52948
Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A (2008) Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA 105:1380–1385
Pauwels L, Inzé D, Goossens A (2009) Jasmoate-inducible gene: what does it mean? Trends Plant Sci 14:87–91
Penninckx IA, Eggermont K, Terras FR, Thomma BP, Samblanx GW, Buchala A, Métraux JP, Manners JM, Broekaert WF (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8:2309–2323
Per TS, Khan MIR, Anjum NA, Masood A, Hussain SJ, Khan NA (2018) Jasmonates in plant under abiotic stresses: crosstalk with other phytohormones matters. Environ Exp Bot 145:104–120
Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, Sharma SB, Klessig DF, Martienssen R, Mattson O, Jensen AB, Mundy J (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103:1111–1120
Pichersky E, Taguso RA (2018) Why do plants produce so many terpenoid compounds? New Phytol 220:692–702
Pieterse CMJ, Van der Does D, Zamioudis C, Leno-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521
Qin Y, Bai S, Li W, Sun T, Galbraith DW, Yang Z, Zhou Y, Sun G, Wang B (2020) Transcriptome analysis reveals key genes involved in the regulation of nicotine biosynthesis at early time points after topping in tobacco (Nicotiana tabacum L). BMC Plant Biol 20:30
Reeves PH, Ellis CM, Ploense SE, Wu MF, Yadav V, Tholl D, Chetelat A, Haupt I, Kennerley BJ, Hodgens C, Farmer EE, Nagpal P, Reed JW (2012) A regulatory network for coordinated flower maturation. PLoS Genet 8:e1002506
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Ru P, Xu L, Ma H, Huang H (2006) Plant fertility defects induced by the enhanced expression of microRNA 167. Cell Res 16:457–465
Shoji T, Hashimoto T (2011) Nicotine biosynthesis in plant metabolism and biotechnology. Wiley, Chichester, pp 191–216
Shoji T, Nakajima K, Hashimoto T (2000) Ethylene suppresses jasmonate-induced gene expression in nicotine biosynthesis. Plant Cell Physiol 60:1072–1076
Shoji T, Kajikawa M, Hashimoto T (2010) Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22:3390–3409
Sibéril Y, Benhamron S, Memelink J, Giglioli-Guivarc’h N, Thiersault M, Boisson B, Doireau P, Gantet P (2001) Catharanthus roseus G-box binding factors 1 and 2 act as repressors of strictosidine synthase gene expression in cell cultures. Plant Mol Biol 45:477–488
Singh SK, Patra B, Paul P, Liu Y, Pattanaik S, Yuan L (2020) Revisiting the ORCA gene cluster that regulates terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Sci 293:110408
Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux J, Brown R, Kazan K, Van Loon LC, Dong X, Pieterse CMJ (2003) NPR1 modulates crosstalk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15:760–770
Sui X, Singh SK, Patra B, Schluttenhofer C, Guo W, Pattanaik S, Yuan L (2018) Cross-family transcription factor interaction between MYC2 and GBFs modulates terpenoid indole alkaloid biosynthesis. J Exp Bot 69:4267–4281
Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, Wu X, Cohen JD, Palme K, Li C (2009) Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21:1495–1511
Tabata R, Ikezaki M, Fujibe T, Aida M, Tian C, Ueno Y, Yamamoto KT, Machida Y, Nakamura K, Ishiguro S (2010) Arabidopsis AUXIN RESPONSE FACTOR6 and 8 regulate jasmonic acid biosynthesis and flora organ development via repression of class 1 KNOX genes. Plant Cell Physiol 51:164–175
Thangavelu M, Belostotsky D, Bevan MW, Flavell RB, Rogers HJ, Lonsdale DM (1993) Partial characterization of the Nicotiana tabacum actin gene family: evidence for pollen-specific expression of one of the gene family members. Mol Gen Genet 240:290–295
Todd AT, Liu E, Polvi SL, Pammett RT, Page JE (2010) A functional genomics screen identifies diverse transcription factors that regulate alkaloid biosynthesis in Nicotiana benthamiana. Plant J 62:589–600
Trapnell C, Williams BA, Pertea G, Motrazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Wang B, Lewis R, Shi J, Song Z, Gao Y, Li W, Chen H, Qu R (2015) Genetic factors for enhancement of nicotine levels in cultivated tobacco. Sci Rep 5:17360
Wastemack C, Strnad M (2017) Jasmonates are signals in the biosynthesis of secondary metabolites—pathways, transcription factors and applied aspects—a brief review. N Biotechnol 48:1–11
Weijers D, Wagner D (2016) Transcriptional responses to the auxin hormone. Annu Rev Plant Biol 67:539–574
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer, New York
Winz RA, Baldwin IT (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata IV Insect-induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiol 125:2189–2202
Yu G, Wang LG, Han Y, He QY (2012) clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287
Yuan C, Lazarowitz SG, Citovsky V (2017) Identification of plasmodesmal localization sequences in proteins in planta. J Vis Exp 126:55301
Zhang H, Hedhili S, Montiel G, Zhang Y, Chatel G, Pré M, Gantet P, Memelink J (2011) The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J 67:61–71
Zhang X, Zhu Z, An F, Hao D, Li P, Song J, Yi C, Guo H (2014) Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 26:1105–1117
Zhou M, Memelink J (2016) Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol Adv 34:441–449
Zhu Z, An F, Feng Y, Li P, Xue L, Jiang AM, Kim Z, To JM, Li TK, Zhang W, Yu X, Dong Q, Chen Z, Seki WQ, Zhou M, Guo JM (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108:12539–12544
Acknowledgements
The authors thank Dr. Ge Bai (Yunnan Academy of Tobacco Agricultural Sciences, China) for critical comments on this manuscript. This research was supported by the Projects of YATAS (Contract No. 2019530000241005 and 2017YN05), and the National Natural Science Foundation of China (Grant No. 31860069) to XS.
Author information
Authors and Affiliations
Contributions
XS designed the study; MH, YG, ZS, CY, CH, and LZ performed experiments; BW, CH, YZ, and CZ analyzed data; XS, ZH, HM, LW, and CZ wrote the paper. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, M., Zhang, H., Wang, B. et al. Transcriptomic analysis provides insights into the AUXIN RESPONSE FACTOR 6-mediated repression of nicotine biosynthesis in tobacco (Nicotiana tabacum L.). Plant Mol Biol 107, 21–36 (2021). https://doi.org/10.1007/s11103-021-01175-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-021-01175-3