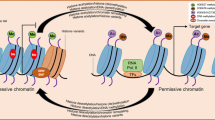
Abstract
C-5 DNA methylation is an essential mechanism controlling gene expression and developmental programs in a variety of organisms. Though the role of DNA methylation has been intensively studied in mammals and Arabidopsis, little is known about the evolution of this mechanism. The chromomethylase (CMT) methyltransferase family is unique to plants and was found to be involved in DNA methylation in Arabidopsis, maize and tobacco. The moss Physcomitrella patens, a model for early terrestrial plants, harbors a single homolog of the CMT protein family designated as PpCMT. Our phylogenetic analysis suggested that the CMT family is unique to embryophytes and its earliest known member PpCMT belongs to the CMT3 subfamily. Thus, P. patens may serve as a model to study the ancient functions of the CMT3 family. We have generated a ΔPpcmt deletion mutant which demonstrated that PpCMT is essential for P. patens protonema and gametophore development and is involved in CHG methylation as demonstrated at four distinct genomic loci. PpCMT protein accumulation pattern correlated with proliferating cells and was sub-localized to the nucleus as predicted from its function. Taken together, our results suggested that CHG DNA methylation mediated by CMT has been employed early in land plant evolution to control developmental programs during both the vegetative and reproductive haploid phases along the plant life cycle.







Similar content being viewed by others
Abbreviations
- BAH:
-
Bromo-adjacent homology domain
- CMT:
-
Chromomethylase
- DNMT:
-
DNA methyltransferase
- DRM:
-
Domains rearranged methyltransferase
- GUS:
-
β-Glucuronidase
- MTD:
-
C-5 DNA methyltransferase domain
- MET1:
-
DNA methyltransferase 1
- RdDM:
-
RNA directed DNA methylation
References
Ashton NW, Cove DJ (1977) The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss Physcomitrella patens. Mol Gen Genet 154:87–95
Bauer MJ, Fischer RL (2011) Genome demethylation and imprinting in the endosperm. Curr Opin Plant Biol 14(2):162–167
Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE (2008) Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE 3(9):e3156
Bezanilla M, Pan A, Quatrano RS (2003) RNA interference in the moss Physcomitrella patens. Plant Physiol 133(2):470–474
Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13(24):2212–2217
Cao X, Jacobsen SE (2002) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99(Suppl 4):16491–16498
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17(4):540–552
Cedar H, Bergman Y (2012) Programming of DNA methylation patterns. Annu Rev Biochem 81:97–117
Chevenet F, Brun C, Banuls AL, Jacq B, Christen R (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439
Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452(7184):215–219
Cove D (2005) The moss Physcomitrella patens. Annu Rev Genet 39:339–358
Deleris A, Stroud H, Bernatavichute Y, Johnson E, Klein G, Schubert D, Jacobsen SE (2012) Loss of the DNA methyltransferase MET1 Induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet 8(11):e1003062
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36(Web Server issue):W465–W469
Ebbs ML, Bender J (2006) Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18(5):1166–1176
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107(19):8689–8694
Feng S, Jacobsen SE (2011) Epigenetic modifications in plants: an evolutionary perspective. Curr Opin Plant Biol 14(2):179–186
Frank W, Decker EL, Reski R (2005) Molecular tools to study Physcomitrella patens. Plant Biol (Stuttg) 7(3):220–227
Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74:481–514
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40(Database issue):D1178–D1186
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52(5):696–704
Henderson IR, Chan SR, Cao X, Johnson L, Jacobsen SE (2010a) Accurate sodium bisulfite sequencing in plants. Epigenetics 5(1):47–49
Henderson IR, Deleris A, Wong W, Zhong X, Chin HG, Horwitz GA, Kelly KA, Pradhan S, Jacobsen SE (2010b) The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet 6(10):e1001182
Henikoff S, Comai L (1998) A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics 149(1):307–318
Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 89(22):10915–10919
Hou PQ, Lee YI, Hsu KT, Lin YT, Wu WZ, Lin JY, Nam TN, Fu SF (2013) Functional characterization of Nicotiana benthamiana chromomethylase 3 in developmental programs by virus-induced gene silencing. Physiol Plant 150(1):119–132
Ishikawa M, Murata T, Sato Y, Nishiyama T, Hiwatashi Y, Imai A, Kimura M, Sugimoto N, Akita A, Oguri Y, Friedman WE, Hasebe M, Kubo M (2011) Physcomitrella cyclin-dependent kinase A links cell cycle reactivation to other cellular changes during reprogramming of leaf cells. Plant Cell 23(8):2924–2938
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6(13):3901–3907
Jullien PE, Berger F (2010) DNA methylation reprogramming during plant sexual reproduction? Trends Genet 26(9):394–399
Jurkowska RZ, Jurkowski TP, Jeltsch A (2011) Structure and function of mammalian DNA methyltransferases. ChemBioChem 12(2):206–222
Kamisugi Y, Schlink K, Rensing SA, Schween G, von Stackelberg M, Cuming AC, Reski R, Cove DJ (2006) The mechanism of gene targeting in Physcomitrella patens: homologous recombination, concatenation and multiple integration. Nucleic Acids Res 34(21):6205–6214
Katoh K, Asimenos G, Toh H (2009) Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 537:39–64
Katz A, Oliva M, Mosquna A, Hakim O, Ohad N (2004) FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J 37(5):707–719
Landberg K, Pederson ER, Viaene T, Bozorg B, Friml J, Jonsson H, Thelander M, Sundberg E (2013) The moss Physcomitrella patens reproductive organ development is highly organized, affected by the two SHI/STY genes and by the level of active auxin in the SHI/STY expression domain. Plant Physiol 162(3):1406–1419.
Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11(3):204–220
Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, Carrington JC, Doerge RW, Colot V, Martienssen R (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430(6998):471–476
Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133(3):523–536
Loytynoja A, Goldman N (2010) webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics 11:579
Malik G, Dangwal M, Kapoor S, Kapoor M (2012) Role of DNA methylation in growth and differentiation in Physcomitrella patens and characterization of cytosine DNA methyltransferases. FEBS J 279(21):4081–4094
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39(Database issue):D225–D229
Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G (2008) Cyclical DNA methylation of a transcriptionally active promoter. Nature 452(7183):45–50
Mosquna A, Katz A, Decker EL, Rensing SA, Reski R, Ohad N (2009) Regulation of stem cell maintenance by the Polycomb protein FIE has been conserved during land plant evolution. Development 136(14):2433–2444
Nishiyama R, Ito M, Yamaguchi Y, Koizumi N, Sano H (2002) A chloroplast-resident DNA methyltransferase is responsible for hypermethylation of chloroplast genes in Chlamydomonas maternal gametes. Proc Natl Acad Sci USA 99(9):5925–5930
Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7(1):9–17
Okano Y, Aono N, Hiwatashi Y, Murata T, Nishiyama T, Ishikawa T, Kubo M, Hasebe M (2009) A polycomb repressive complex 2 gene regulates apogamy and gives evolutionary insights into early land plant evolution. Proc Natl Acad Sci USA 106(38):16321–16326
Papa CM, Springer NM, Muszynski MG, Meeley R, Kaeppler SM (2001) Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell 13(8):1919–1928
Pavlopoulou A, Kossida S (2007) Plant cytosine-5 DNA methyltransferases: structure, function, and molecular evolution. Genomics 90(4):530–541
Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, Tanahashi T, Sakakibara K, Fujita T, Oishi K, Shin IT, Kuroki Y, Toyoda A, Suzuki Y, Hashimoto S, Yamaguchi K, Sugano S, Kohara Y, Fujiyama A, Anterola A, Aoki S, Ashton N, Barbazuk WB, Barker E, Bennetzen JL, Blankenship R, Cho SH, Dutcher SK, Estelle M, Fawcett JA, Gundlach H, Hanada K, Heyl A, Hicks KA, Hughes J, Lohr M, Mayer K, Melkozernov A, Murata T, Nelson DR, Pils B, Prigge M, Reiss B, Renner T, Rombauts S, Rushton PJ, Sanderfoot A, Schween G, Shiu SH, Stueber K, Theodoulou FL, Tu H, Van de Peer Y, Verrier PJ, Waters E, Wood A, Yang L, Cove D, Cuming AC, Hasebe M, Lucas S, Mishler BD, Reski R, Grigoriev IV, Quatrano RS, Boore JL (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319(5859):64–69
Sakakibara K, Nishiyama T, Deguchi H, Hasebe M (2008) Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol Dev 10(5):555–566
Sakakibara K, Nishiyama T, Kato M, Hasebe M (2001) Isolation of homeodomain-leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain-leucine zipper genes in land plants. Mol Biol Evol 18(4):491–502
Saze H, Kakutani T (2011) Differentiation of epigenetic modifications between transposons and genes. Curr Opin Plant Biol 14(1):81–87
Schaefer M, Lyko F (2010) Solving the Dnmt2 enigma. Chromosoma 119(1):35–40
Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152(1–2):352–364
Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153(1):193–205
Zemach A, McDaniel IE, Silva P, Zilberman D (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328(5980):916–919
Zemach A, Zilberman D (2010) Evolution of eukaryotic DNA methylation and the pursuit of safer sex. Curr Biol 20(17):R780–R785
Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126(6):1189–1201
Zimmer AD, Lang D, Buchta K, Rombauts S, Nishiyama T, Hasebe M, Van de Peer Y, Rensing SA, Reski R (2013) Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genom 14:498
Acknowledgments
We thank Professors Tal Pupko, Eran Bacharach, Shaul Yalovsky and Daniel Chamovitz for critical reading and helpful comments. C.N.M and R.Y were supported in part by a matching Tel-Aviv University Deans doctoral fellowship and the Manna foundation. This research was supported by the Israeli Science Foundation Grant #767/09, and by the Israel Korea program # 3-824 financed by the Ministry of Science and Technology, both granted to N.O. Transgenic lines described in this study were deposited in the International Moss Stock Center (http://www.moss-stock-center.org/) with the accessions IMSC 40736, 40737, 40738 (ΔPpcmt 103, 157 and 281) and 40739, 40740 (PpCMT–GUS 7 and 8).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Chen Noy-Malka and Rafael Yaari have contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Noy-Malka, C., Yaari, R., Itzhaki, R. et al. A single CMT methyltransferase homolog is involved in CHG DNA methylation and development of Physcomitrella patens . Plant Mol Biol 84, 719–735 (2014). https://doi.org/10.1007/s11103-013-0165-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0165-6