Abstract
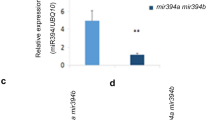
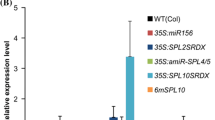
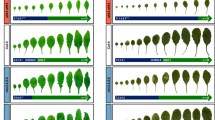
In plants, developmental timing is coordinately regulated by a complex signaling network that integrates diverse intrinsic and extrinsic signals. miR172 promotes photoperiodic flowering. It also regulates adult development along with miR156, although the molecular mechanisms underlying this regulation are not fully understood. Here, we demonstrate that miR172 modulates the developmental transitions by regulating the expression of a subset of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes, which are also regulated by miR156. The SPL3/4/5 genes were upregulated in the miR172-overproducing plants (35S:172) and its target gene mutants that exhibit early flowering. In contrast, expression of other SPL genes was not altered to a discernible level. Kinetic measurements of miR172 abundance in the transgenic plants expressing the MIR156a gene driven by a β-estradiol-inducible promoter revealed that expressions of miR172 and miR156 are not directly interrelated. Instead, the 2 miRNA signals are integrated at the SPL3/4/5 genes. Notably, analysis of developmental patterns in the 156 × 172 plants overproducing both miR172 and miR156 showed that whereas vegetative phase change was delayed as observed in the miR156-overproducing plants (35S:156), flowering initiation was accelerated as observed in the 35S:172 transgenic plants. Together, these observations indicate that although miR172 and miR156 play distinct roles in the timing of developmental phase transitions, there is a signaling crosstalk mediated by the SPL3/4/5 genes.







Similar content being viewed by others
Abbreviations
- AP2:
-
APETALA 2
- Cg1:
-
Corngrass1
- FT:
-
FLOWERING LOCUS T
- Gl15:
-
Glossy15
- GUS:
-
β-Glucuronidase
- LD:
-
Long day
- miRNA:
-
MicroRNA
- qRT-PCR:
-
Quantitative real-time RT-PCR
- RT-PCR:
-
Reverse transcription-PCR
- SD:
-
Short day
- SMZ:
-
SCHLAFMÜTZE
- SNZ:
-
SCHNARCHZAPFEN
- SOC1:
-
SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1
- SPL3/4/5:
-
SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3/4/5
- TOE1/2:
-
TARGET OF EAT 1/2
References
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Bäurle I, Dean C (2006) The timing of developmental transitions in plants. Cell 125:655–664
Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS (2003) HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130:1493–1504
Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13:1499–1510
Cardon GH, Höhmann S, Nettesheim K, Saedler H, Huijser P (1997) Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J 12:367–377
Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022–2025
Chen J, Li WX, Xie D, Peng JR, Ding SW (2004) Viral virulence protein suppresses RNA silencing–mediated defense but upregulates the role of microRNA in host Gene expression. Plant Cell 16:1302–1313
Chuck G, Cigan AM, Saeteurn K, Hake S (2007) The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet 39:544–549
Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49:683–693
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618
Hunter C, Sun H, Poethig RS (2003) The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol 13:1734–1739
Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19:2736–2748
Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, Kim JA, Lee M, Lee Y, Narry Kim V, Chua NH, Park CM (2005) microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J 42:84–94
Kim YS, Kim SG, Lee M, Lee I, Park HY, Seo PJ, Jung JH, Kwon EJ, Suh SW, Paek KH, Park CM (2008) HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20:920–933
Lauter N, Kampani A, Carlson S, Goebel M, Moose SP (2005) microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA 102:9412–9417
Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Kim J, Hong CB, Kim HJ, Park CM (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16:731–740
Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLOS Biol 7:e1000148
Moose SP, Sisco PH (1996) glossy15, an APETELA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev 10:3018–3027
Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18:2368–2379
Poethig RS (2003) Phase change and the regulation of developmental timing in plants. Science 301:334–336
Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU (2003) Dissection of floral induction pathways using global expression analysis. Development 130:6001–6012
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P (2008) The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol 67:183–195
Scott DB, Jin W, Ledford HK, Jung HS, Honma MA (1999) EAF1 regulates vegetative-phase change and flowering time in Arabidopsis. Plant Physiol 120:675–684
Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124:645–654
Telfer A, Poethig RS (1998) HASTY: a gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125:1889–1898
Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16:69–79
Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20:1231–1243
Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138:738–749
Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133:3539–3547
Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759
Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D (2009) The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17:268–278
Acknowledgments
We thank Narry Kim for scientific discussions and Emily Wheeler for editorial assistance. This work was supported by the Brain Korea 21 and Biogreen 21 (20080401034001) Programs and by grants from the Plant Signaling Network Research Center, the National Research Foundation of Korea (20100014373 and 2009-0087317), and from the Agricultural R & D Promotion Center (309017-5), Korea Ministry for Food, Agriculture, Forestry and Fisheries.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jae-Hoon Jung and Pil Joon Seo have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jung, JH., Seo, P.J., Kang, S.K. et al. miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol Biol 76, 35–45 (2011). https://doi.org/10.1007/s11103-011-9759-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-011-9759-z