Abstract
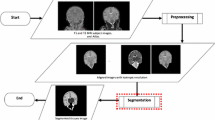
Brain extraction is one of the most important preprocessing steps in cerebral magnetic resonance (MR) image analysis. Brain extraction from neonatal MR images is particularly challenging due to significant differences in head size and shape between neonates and rapid changes in neonatal brain structure in the weeks and months after birth. In this work, a multi-atlas-based neonatal brain extraction method using atlas library clustering and local label fusion (NOBELL) is presented. In NOBELL, an affinity propagation (AP) approach is first applied to cluster images of an atlas library into clusters represented by exemplars, which are used to select best matching clusters for target images. A local weighted voting strategy based on Jacobian determinant ranking is then employed to extract brain from target images using training images in best matching clusters. The performance of NOBELL was evaluated on T2- and T1-weighted scans of 40 neonates aged between 37 and 44 weeks. NOBELL outperformed two popular brain extraction tools, FSL’s Brain Extraction Tool (BET) and BrainSuite’s Brain Surface Extractor (BSE), and achieved higher accuracy with brain masks very close to manually extracted ones. NOBELL showed an average Jaccard coefficient of 0.974 (0.942) on T2 (T1)-weighted images in comparison with 0.908 (0.602) and 0.845 (0.762) achieved by BSE, and BET, respectively. NOBELL allows for accurate and efficient brain extraction, a crucial step in brain MRI applications such as accurate brain tissue segmentation and volume estimation as well as accurate cortical surface delineation in neonates.









Similar content being viewed by others
References
Aboutanos GB, Nikanne J, Watkins N, Dawan BM (1999) Model creation and deformation for the automatic segmentation of the brain in MR images. IEEE Trans Biomed Eng 46(11):1346–1356. https://doi.org/10.1109/10.797995
Alansary A, Ismail M, Soliman A, Khalifa F, Nitzken M, Elnakib A, Mostapha M, Black A, Stinebruner K, Casanova MF, Zurada JM, El-Baz A (2016) Infant brain extraction in T1-weighted MR images using BET and refinement using LCDG and MGRF models. IEEE Journal of Biomedical and Health Informatics 20(3):925–935. https://doi.org/10.1109/jbhi.2015.2415477
Andermatt S, Pezold S, Cattin P (2016) Multi-dimensional gated recurrent units for the segmentation of biomedical 3D-data. Deep Learning and Data Labeling for Medical Applications Springer https://doi.org/10.1007/978-3-319-46976-8_15
Ashburner J, Barnes G, Chen C, Daunizeau J, Flandin G, Friston K, Kiebel S, Kilner J, Litvak V, Moran R (2014) Wellcome Trust Centre for Neuroimaging. Wellcome Trust, London
Brummer ME, Mersereau RM, Eisner RL, Lewine RRJ (1993) Automatic detection of brain contours in MRI data sets. IEEE Trans Med Imaging 12(2):153–166. https://doi.org/10.1109/42.232244
Chang HH, Zhuang AH, Valentino DJ, Chu WC (2009) Performance measure characterization for evaluating neuroimage segmentation algorithms. NeuroImage 47(1):122–135. https://doi.org/10.1016/j.neuroimage.2009.03.068
Chiverton J, Wells K, Lewis E, Chen C, Podda B, Johnson D (2007) Statistical morphological skull stripping of adult and infant MRI data. Comput Biol Med 37(3):342–357. https://doi.org/10.1016/j.compbiomed.2006.04.001
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26(3):297–302. https://doi.org/10.2307/1932409
Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C (2013) Multi-atlas skull-stripping. Acad Radiol 20(12):1566–1576. https://doi.org/10.1016/j.acra.2013.09.010
Fonov V, Evans A, McKinstry R, Almli CR, Collins L (2009) Unbiased nonlinear average age-appropriate brain templates from birth to adulthood Nueroimage:47. https://doi.org/10.1016/S1053-8119(09)70884-5
Frey BJ, Dueck D (2007) Clustering by passing messages between data points. Science 315(5814):972–976. https://doi.org/10.1126/science.1136800
Galdames FJ, Jaillet F, Perez CA (2012) An accurate skull stripping method based on simplex meshes and histogram analysis for magnetic resonance images. J Neurosci Methods 206(2):103–119. https://doi.org/10.1016/j.jneumeth.2012.02.017
Gao J, Xie M (2009) Skull-stripping MR brain images using anisotropic diffusion filtering and morphological processing. Paper presented at the International Symposium on Computer Network and Multimedia Technology,
Grau V, Mewes A, Alcaniz M, Kikinis R, Warfield SK (2004) Improved watershed transform for medical image segmentation using prior information. IEEE Trans Med Imaging 23(4):447–458. https://doi.org/10.1109/TMI.2004.824224
Hahn HK, Peitgen HO (2000) The skull stripping problem in MRI solved by a single 3D watershed transform. Paper presented at the International Conference on Medical Image Computing and Computer-Assisted Intervention,
Heckemann RA, Ledig C, Gray KR, Aljabar P, Rueckert D, Hajnal JV, Hammers A (2015) Correction: brain extraction using label propagation and group agreement: pincram. PLoS One 10(8):e0135746. https://doi.org/10.1371/journal.pone.0135746
Hughes EJ, Winchman T, Padormo F, Teixeira R, Wurie J, Sharma M, Fox M, Hutter J, Cordero-Grande L, Price AN (2017) A dedicated neonatal nrain imaging system. Magn Reson Med 78(2):794–804. https://doi.org/10.1002/mrm.26462
Hwang J, Han Y, Park H (2011) Skull-stripping method for brain MRI using a 3D level set with a speedup operator. J Magn Reson Imaging 34(2):445–456. https://doi.org/10.1002/jmri.22661
Jaccard P (1912) The distribution of the flora in the alpine zone. New Phytol 11(2):37–50. https://doi.org/10.1111/j.1469-8137.1912.tb05611.x
Jenkinson M, Pechaud M, Smith SM (2005) BET2: MR-based estimation of brain, skull and scalp surfaces. Paper presented at the Eleventh Annual Meeting of the Organization for Human Brain Mapping,
Kalavathi P (2014) Computation of brain asymmetry in 2D MR brain images. Int J Sci Eng Res 5 (7)
Kalavathi P, Prasath VB (2016) Methods on skull stripping of MRI head scan images-a review. J Digit Imaging 29(3):365–379. https://doi.org/10.1007/s10278-015-9847-8
Kleesiek J, Urban G, Hubert A, Schwarz D, Maier-Hein K, Bendszus M, Biller A (2016) Deep MRI brain extraction: a 3D convolutional neural network for skull stripping. NeuroImage 129:460–469. https://doi.org/10.1016/j.neuroimage.2016.01.024
Kobashi S, Fujimoto Y, Ogawa M, Ando K, Ishikura R, Kondo K, Hirota S, Hata Y (2007) Fuzzy-ASM based automated skull stripping method from infantile brain MR images. Paper presented at the IEEE International Conference on Granular Computing,
Lemieux L, Hagemann G, Krakow K, Woermann FG (1999) Fast automatic segmentation of the brain in T1-weighted volume MRI data. Paper presented at the SPIE Conference on Image Processing,
Leung KK, Barnes J, Modat M, Ridgway GR, Bartlett JW, Fox NC, Ourselin S (2011) Brain MAPS: an automated, accurate and robust brain extraction technique using a template library. NeuroImage 55(3):1091–1108. https://doi.org/10.1016/j.neuroimage.2010.12.067
Liu JX, Chen YS, Chen LF (2009) Accurate and robust extraction of brain regions using a deformable model based on radial basis functions. J Neurosci Methods 183(2):255–266. https://doi.org/10.1016/j.jneumeth.2009.05.011
Makropoulos A, Gousias IS, Ledig C, Aljabar P, Serag A, Hajnal JV, Edwards AD, Counsell SJ, Rueckert D (2014) Automatic whole brain MRI segmentation of the developing neonatal brain. IEEE Trans Med Imaging 33(9):1818–1831. https://doi.org/10.1109/TMI.2014.2322280
Makropoulos A, Robinson EC, Schuh A, Wright R, Fitzgibbon S, Bozek J, Counsell SJ, Steinweg J, Vecchiato K, Passerat-Palmbach J (2018) The developing human connectome project: a minimal processing pipeline for neonatal cortical surface reconstruction. NeuroImage 173:88–112. https://doi.org/10.1016/j.neuroimage.2018.01.054
Noorizadeh N, Kazemi K, Danyali H, Aarabi A (2019) Multi-atlas based neonatal brain extraction using a two-level patch-based label fusion strategy. Biomedical Signal Processing and Control 54:101602. https://doi.org/10.1016/j.bspc.2019.101602
Park JG, Lee C (2009) Skull stripping based on region growing for magnetic resonance brain images. NeuroImage 47(4):1394–1407. https://doi.org/10.1016/j.neuroimage.2009.04.047
Péporté M, Ghita DEI, Twomey E, Whelan PF (2011) A hybrid approach to brain extraction from premature infant MRI. Paper presented at the Scandinavian Conference on Image Analysis,
Rousseau F, Habas PA, Studholme C (2011) A supervised patch-based approach for human brain labeling. IEEE Trans Med Imaging 30(10):1852–1862. https://doi.org/10.1109/TMI.2011.2156806
Sadananthan SA, Zheng W, Chee MW, Zagorodnov V (2010) Skull stripping using graph cuts. NeuroImage 49(1):225–239. https://doi.org/10.1016/j.neuroimage.2009.08.050
Sandor S, Leahy R (1997) Surface-based labeling of cortical anatomy using a deformable atlas. IEEE Trans Med Imaging 16(1):41–54. https://doi.org/10.1109/42.552054
Serag A, Blesa M, Moore EJ, Pataky R, Sparrow SA, Wilkinson A, Macnaught G, Semple SI, Boardman JP (2016) Accurate learning with few atlases (ALFA): an algorithm for MRI neonatal brain extraction and comparison with 11 publicly available methods. Sci Rep 6:23470. https://doi.org/10.1038/srep23470
Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM (2001) Magnetic resonance image tissue classification using a partial volume model. NeuroImage 13(5):856–876. https://doi.org/10.1006/nimg.2000.0730
Shi F, Wang L, Dai Y, Gilmore JH, Lin W, Shen D (2012) LABEL: pediatric brain extraction using learning-based meta-algorithm. NeuroImage 62(3):1975–1986. https://doi.org/10.1016/j.neuroimage.2012.05.042
Smith SM (2002) Fast robust automated brain extraction. Hum Brain Mapp 17(3):143–155. https://doi.org/10.1002/hbm.10062
Somasundaram K, Kalaiselvi T (2010a) Fully automatic brain extraction algorithm for axial T2-weighted magnetic resonance images. Comput Biol Med 40(10):811–822. https://doi.org/10.1016/j.compbiomed.2010.08.004
Somasundaram K, Kalavathi P (2010b) Automatic skull stripping of magnetic resonance images (MRI) of human head scans using image contour. Paper presented at the The National Conference on Image Processing,
Somasundaram K, Kalavathi P (2010c) A hybrid method for automatic skull stripping of magnetic resonance images (MRI) of human head scans. Paper presented at the international conference on computing communication and networking technologies (ICCCNT),
Somasundaram K, Kalavathi P (2011) Skull stripping of MRI head scans based on 2D region growing. Paper presented at the Proc. ICOM11 Tiruchirappalli, Tamil Nadu,
Tripathi S, Anand R, Fernandez E (2018) A review of brain MR image segmentation techniques. Paper presented at the Proceedings of International Conference on Recent Innovations in Applied Science, Engineering & Technology,
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29(6):1310–1320. https://doi.org/10.1109/TMI.2010.2046908
Valente J, Vieira PM, Couto C, Lima CS (2018) Brain extraction in partial volumes T2*@ 7T by using a quasi-anatomic segmentation with bias field correction. J Neurosci Methods 295:129–138. https://doi.org/10.1016/j.jneumeth.2017.12.006
van Opbroek A, van der Lijn F, de Bruijne M (2013) Automated brain-tissue segmentation by multi-feature SVM classification. Paper presented at the the MICCAI grand challenge on MR brain image segmentation (MRBrainS13),
Wang Y, Nie J, Yap PT, Shi F, Guo L, Shen D (2011) Robust deformable-surface-based skull-stripping for large-scale studies. Paper presented at the International Conference on Medical Image Computing and Computer-Assisted Intervention,
Yu H, He F, Pan Y (2018) A novel region-based active contour model via local patch similarity measure for image segmentation. Multimed Tools Appl 77(18):24097–24119. https://doi.org/10.1007/s11042-018-5697-y
Yu H, He F, Pan Y (2019) A novel segmentation model for medical images with intensity inhomogeneity based on adaptive perturbation. Multimed Tools Appl 78(9):11779–11798. https://doi.org/10.1007/s11042-018-6735-5
Yunjie C, Jianwei Z, Shunfeng W (2009) A new fast chinese visible human brain skull stripping method. Paper presented at the International Conference on Information and Automation,
Zabihzadeh M, Pishghadam M, Kazemi K, Nekooi S, Tahmasebi Birgani MJ, Seilanian-Toosi F (2017) Comparison the accuracy of fetal brain extraction from T2-half-fourier acquisition single-shot turbo spin-echo (HASTE) MRimage with T2-true fast imaging with steady state free precession (TRUFI) MR image by level set algorithm. International journal of pediatrics 5 (3):4561-4567. Doi:https://doi.org/10.22038/IJP.2017.22292.1864
Acknowledgments
This work was supported by the Cognitive Science and Technology Council (CSTC) of Iran under grant numbers 1896, 3308 and by Le Bonheur Children’s Hospital, the Children’s Foundation Research Institute, and the Le Bonheur Associate Board, Memphis, TN.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Noorizadeh, N., Kazemi, K., Danyali, H. et al. Multi-atlas based neonatal brain extraction using atlas library clustering and local label fusion. Multimed Tools Appl 79, 19411–19433 (2020). https://doi.org/10.1007/s11042-020-08749-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-08749-1