Abstract
Background
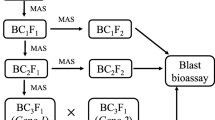
Kashmir valley, India is a homeland to rice landraces like Zag, Nunbeoul, Qadirbeigh, Kawkadur, Kamad, Mushk Budji, etc., generally characterized by short grains, aroma, earliness and cold tolerance. Mushk Budji is a commercially important speciality rice known for its taste and aroma, nonetheless, is extremely vulnerable to blast disease. Through the use of the marker-assisted backcrossing (MABC) approach, a set of 24 Near-isogenic lines (NILs) was created, and the lines with the highest background genome recovery were chosen. The expression analysis was carried out for the component genes and other eight pathway genes related to blast resistance.
Results
The major blast resistance genes Pi9 (from IRBL-9W) and Pi54 (from DHMAS 70Q 164-1b) were incorporated following simultaneous-but-step-wise MABC. The NILs harbouring genes Pi9 + Pi54, Pi9 and Pi54 expressed resistance to isolate (Mo-nwi-kash-32) under controlled and natural field conditions. The loci controlling ETI (effector triggered immunity) included the gene Pi9 and showed 61.18 and 60.27 fold change in relative gene expression in Pi54 + Pi9 and Pi9 carrying NILs against RP Mushk Budji. Pi54 was up regulated and showed 41 and 21 fold change in relative gene expression for NIL-Pi54 + Pi9 and NIL-Pi54, respectively. Among the pathway genes, LOC_Os01g60600 (WRKY 108) recorded 8 and 7.5 fold up regulation in Pi9 and Pi54 NILs.
Conclusion
The NILs showed recurrent parent genome recovery (RPG) per cent of 81.67 to 92.54 and were on par in performance to recurrent parent Mushk Budji. The lines were utilized to study the expression of the loci controlling WRKYs, peroxidases and chitinases that confer overall ETI response.





Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article (and its supplementary information files).
References
Silué D, Notteghem JL, Tharreau D (1992) Evidence of a gene-for-gene relationship in the Oryza sativa–Magnaporthe grisea pathosystem. Phytopathology 82(5):577–580
Brar DS, Khush GS (2018) Wild relatives of rice: a valuable genetic resource for genomics and breeding research. The wild Oryza genomes. Springer, Cham, pp 1–25
Talbot NJ (2003) On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu Rev Microbiol 57(1):177–202
Khanna A, Sharma V, Ellur RK, Shikari AB, Krishnan SG, Singh U et al (2015) Marker assisted pyramiding of major blast resistance genes Pi9 and Pita in the genetic background of an elite Basmati rice variety, Pusa Basmati 1. Indian J Genet 75(4):417–425
Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L et al (2006) The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172(3):1901–1914
Sharma T, Madhav M, Singh B, Shanker P, Jana T, Dalal V et al (2005) High-resolution mapping, cloning and molecular characterization of the Pi-k h gene of rice, which confers resistance to Magnaporthe grisea. Mol Genet Genomics 274(6):569–578
Rai AK, Kumar SP, Gupta SK, Gautam N, Singh NK, Sharma TR (2011) Functional complementation of rice blast resistance gene Pi-k h (Pi54) conferring resistance to diverse strains of Magnaporthe oryzae. J Plant Biochem Biotechnol 20(1):55–65
Gupta SK, Rai AK, Kanwar SS, Chand D, Singh NK, Sharma TR (2012) The single functional blast resistance gene Pi54 activates a complex defence mechanism in rice. J Exp Bot 63(2):757–772
Jones JD, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329
Khan GH, Shikari AB, Vaishnavi R, Najeeb S, Padder BA, Bhat ZA et al (2018) Marker-assisted introgression of three dominant blast resistance genes into an aromatic rice cultivar Mushk Budji. Sci Rep 8(1):1–13
Shikari AB, Najeeb S, Khan G, Mohidin FA, Shah AH, Nehvi FA et al (2021) KASP™ based markers reveal a population sub-structure in temperate rice (Oryza sativa L.) germplasm and local landraces grown in the Kashmir valley, north-western Himalayas. Genetic Resour Crop Evol 68(3):821–34
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M et al (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9(6):199–207
van Berloo R, Aalbers H, Werkman A, Niks RE (2001) Resistance QTL confirmed through development of QTL-NILs for barley leaf rust resistance. Mol Breed 8(3):187–195
Murray M, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8(19):4321–4326
Bonman J, Vergel de Dios T, Khin M (1986) Physiologic specialization of Pyricularia oryzae in the Philippines. Plant Dis 70(8):767–769
Mackill D, Bonman J (1992) Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82(7):746–749
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Hittalmani S, Parco A, Mew T, Zeigler R, Huang N (2000) Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor Appl Genet 100(7):1121–1128
Ramkumar G, Srinivasarao K, Mohan KM, Sudarshan I, Sivaranjani A, Gopalakrishna K et al (2011) Development and validation of functional marker targeting an InDel in the major rice blast disease resistance gene Pi54 (Pik h). Mol Breed 27(1):129–135
Tanksley SD, Young ND, Paterson AH, Bonierbale M (1989) RFLP mapping in plant breeding: new tools for an old science. Bio/Technology 7(3):257–264
Hospital F, Chevalet C, Mulsant P (1992) Using markers in gene introgression breeding programs. Genetics 132(4):1199–1210
Cruz ND, Khush G (2000) Rice grain quality evaluation procedures. Aromatic Rices 3:15–28
Shikari AB, Khanna A, Krishnan SG, Singh U, Rathour R, Tonapi V et al (2013) Molecular analysis and phenotypic validation of blast resistance genes Pita and Pita2 in landraces of rice (Oryza sativa L.). Indian J Genet 73(2):131–14
Shikari AB, Rajashekara H, Khanna A, Krishnan SG, Rathour R, Singh U et al (2014) Identification and validation of rice blast resistance genes in Indian rice germplasm. Indian J Genet Plant Breed 74(3):286–299
Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H et al (1999) The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J 19(1):55–64
Bryan GT, Wu K-S, Farrall L, Jia Y, Hershey HP, McAdams SA et al (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12(11):2033–2045
Sharma T, Rai A, Gupta S, Singh N (2010) Broad-spectrum blast resistance gene Pi-k h cloned from rice line Tetep designated as Pi54. J Plant Biochem Biotechnol 19(1):87–89
Sharma T, Shanker P, Singh B, Jana T, Madhav M, Gaikwad K et al (2005) Molecular mapping of rice blast resistance gene Pi-k h in the rice variety Tetep. J Plant Biochem Biotechnol 14(2):127–133
Singh A, Gopalakrishnan S, Singh V, Prabhu K, Mohapatra T, Singh N et al (2011) Marker assisted selection: a paradigm shift in Basmati breeding. Indian J Genet Plant Breed 71(2):120
Karmakar S, Molla KA, Das K, Sarkar SN, Datta SK, Datta K (2017) Dual gene expression cassette is superior than single gene cassette for enhancing sheath blight tolerance in transgenic rice. Sci Rep 7:7900. https://doi.org/10.1038/s41598-017-08180-x.chitinase
Lee S-K, Song M-Y, Seo Y-S, Kim H-K, Ko S, Cao P-J et al (2009) Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil–nucleotide-binding–leucine-rich repeat genes. Genetics 181(4):1627–1638
Beffa RS, Hofer R-M, Thomas M, Meins F Jr (1996) Decreased susceptibility to viral disease of [beta]-1, 3-glucanase-deficient plants generated by antisense transformation. Plant Cell 8(6):1001–1011
Hammond-Kosack KE, Jones J (1996) Resistance gene-dependent plant defense responses. Plant Cell 8(10):1773
Imam J, Alam S, Mandal NP, Variar M, Shukla P (2014) Molecular screening for identification of blast resistance genes in North East and Eastern Indian rice germplasm (Oryza sativa L.) with PCR based makers. Euphytica. 196(2):199–211
Variar M, Cruz CV, Carrillo M, Bhatt J, Sangar R (2009) Rice blast in India and strategies to develop durably resistant cultivars. In: Advances in genetics, genomics and control of rice blast disease: Springer, Dordrecht, pp 359–373
Jain P, Singh PK, Kapoor R, Khanna A, Solanke AU, Krishnan SG et al (2017) Understanding host-pathogen interactions with expression profiling of NILs carrying rice-blast resistance Pi9 gene. Front Plant Sci 8:93
Eulgem T (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10(2):71–78
López-Berges MS, Rispail N, Prados-Rosales RC, Di Pietro A (2010) A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell 22(7):2459–2475
Liu J, Liu X, Dai L, Wang G (2007) Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J Genet Genomics 34(9):765–776
Guo S, Boyd J, Sammynaiken R, Loewen MC (2008) Identification and characterization of a unique cysteine residue proximal to the catalytic site of Arabidopsis thaliana carotenoid cleavage enzyme 1. Biochem Cell Biol 86(3):262–270
Quiroga M, Guerrero C, Botella MA, Barceló A, Amaya I, Medina MI et al (2000) A tomato peroxidase involved in the synthesis of lignin and suberin. Plant Physiol 122(4):1119–1128
Almagro L, Gómez Ros L, Belchi-Navarro S, Bru R, Ros Barceló A, Pedreño M (2009) Class III peroxidases in plant defence reactions. J Exp Bot 60(2):377–390
Marjamaa K, Kukkola EM, Fagerstedt KV (2009) The role of xylem class III peroxidases in lignification. J Exp Bot 60(2):367–376
Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K et al (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9(4):436–442
Ton J, Flors V, Mauch-Mani B (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci 14(6):310–317
Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V et al (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141(2):436–445
Montillet J-L, Chamnongpol S, Rustérucci C, Dat J, Van De Cotte B, Agnel J-P et al (2005) Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol 138(3):1516–1526
Wally O, Punja Z (2010) Enhanced disease resistance in transgenic carrot (Daucus carota L.) plants over-expressing a rice cationic peroxidase. Planta 232(5):1229–39
Mauch F, Hadwiger LA, Boller T (1988) Antifungal hydrolases in pea tissue: I. Purification and characterization of two chitinases and two β-1,3-glucanases differentially regulated during development and in response to fungal infection. Plant Physiol 87(2):325–33
Woloshuk CP, Meulenhoff JS, Sela-Buurlage M, Van den Elzen P, Cornelissen B (1991) Pathogen-induced proteins with inhibitory activity toward Phytophthora infestans. Plant Cell 3(6):619–628
Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melchers LS, van den Elzen PJ, Cornelissen BJ (1993) Only specific tobacco (Nicotiana tabacum) chitinases and β-1,3-glucanases exhibit antifungal activity. Plant Physiol 101(3):857–863
Brogue K, Chet I, Holliday M, Cressman R, Biddle P, Knowlton S et al (1991) Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254(5035):1194–1197
Zhu Q, Maher EA, Masoud S, Dixon RA, Lamb CJ (1994) Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase genes in transgenic tobacco. Bio/Technology 12(8):807–812
Lin W, Anuratha C, Datta K, Potrykus I, Muthukrishnan S, Datta SK (1995) Genetic engineering of rice for resistance to sheath blight. Bio/Technology 13(7):686–691
Nishizawa Y, Nishio Z, Nakazono K, Soma M, Nakajima E, Ugaki M et al (1999) Enhanced resistance to blast (Magnaporthe grisea) in transgenic Japonica rice by constitutive expression of rice chitinase. Theor Appl Genet 99(3–4):383–390
Datta K, Tu J, Oliva N, Ona I, Velazhahan R, Mew TW et al (2001) Enhanced resistance to sheath blight by constitutive expression of infection-related rice chitinase in transgenic elite indica rice cultivars. Plant Sci 160(3):405–414
Kasprzewska A (2003) Plant chitinases-regulation and function. Cell Mol Biol Lett 8(3):809–824
McGee JD, Hamer JE, Hodges TK (2001) Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea. Mol Plant Microbe Interact 14(7):877–886
Kim ST, Kim SG, Hwang DH, Kang SY, Kim HJ, Lee BH et al (2004) Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics 4(11):3569–3578
Kim ST, Yu S, Kang YH, Kim SG, Kim J-Y, Kim S-H et al (2008) The rice pathogen-related protein 10 (JIOsPR10) is induced by abiotic and biotic stresses and exhibits ribonuclease activity. Plant Cell Rep 27(3):593–603
Li W, Liu Y, Wang J, He M, Zhou X, Yang C et al (2016) The durably resistant rice cultivar D igu activates defence gene expression before the full maturation of Magnaporthe oryzae appressorium. Mol Plant Pathol 17(3):354–368
Jain P, Dubey H, Singh PK, Solanki AU, Singh AK, Sharma TR (2019) Deciphering signalling network in broad spectrum Near Isogenic Lines of rice resistant to Magnaporthe oryzae. Sci Rep 9:16939. https://doi.org/10.1038/s41598-019-50990-8
Tabien RE, Li Z, Paterson AH, Marchetti MA, Stansel JW, Pinson SRM, Park WD (2000) Mapping of four major rice blast resistance genes from ‘Lemont’and ‘Teqing’ and evaluation of their combinatorial effect for field resistance. Theor Appl Genet 101:1215–1225
Kiryowa M, Nkalubo ST, Mukankusi C, Male A, Gibson P, Tukamuhabwa P, Rubaihayo P (2021) Effectiveness of pyramided genes in conferring resistance to nthracnose disease in common bean populations. Journal of Plant Breeding and Crop Science 13(1):1–13
Divya D, Madhavi KR, Dass MA et al (2018) Expression profile of defense genes in rice lines pyramided with resistance genes against bacterial blight, fungal blast and insect gall midge. Rice 11:40. https://doi.org/10.1186/s12284-018-0231-4
Xiao W, Luo L, Wang H, Guo T, Liu Y, Zhou J, Zhu X, Yang Q, Chen Z (2016) Pyramiding of Pi46 and Pita to improve blast resistance and to evaluate the resistance effect of the two R genes. J Integr Agric 15:2290–2298. https://doi.org/10.1016/S2095-3119(16)61415-6
Yu MM, Dai ZY, Pan CH, Chen XJ, Yu L, Zhang XX, Li YH, Xiao N, Gong HB, Sheng SL, Pan XB, Zhang HX, Li AH (2013) Resistance spectrum difference between two broad- spectrum blast resistance genes, Pigm and Pi2, and their interaction effect on Pi1. Acta Agron Sin 39(11):1927–1934
Shikari AB, Hussain SZ, Parray G, Rather A, Wani SA (2008) Physico-chemical and cooking properties of non/basmati temperate rice (Oryza sativa L.). Crop Improvement 35(2):109
Acknowledgements
The authors acknowledge SKUAST-K for providing Institutional support to carry out the research.
Funding
Part of the work was carried out with the help of financial assistance received from Department of Biotechnology, Govt. of India granted under Project Code: BT/PR8530/AGII/106/935/2014 and is highly acknowledged.
Author information
Authors and Affiliations
Contributions
ABS conceptualized the plan of research work. AS, SM and ABS carried out the marker-assisted foreground and background analysis of NILs. AS, KZ and ABS carried out the expression analysis. SN, GK, RK and JL raised the backcross populations in field and performed crossing work. FAM and ABS prepared the fungus cultures and performed controlled disease phenotyping process. AS and ABS mainly wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shafi, A., Khan, R.S., Mir, S. et al. Gene expression of near-isogenic lines (NILs) carrying blast resistance genes Pi9 and Pi54 in the background of rice cultivar Mushk Budji. Mol Biol Rep 50, 5901–5915 (2023). https://doi.org/10.1007/s11033-023-08475-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08475-5