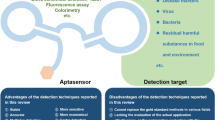
Abstract
Since cells are the basic structural and functional units of organisms, the detection or quantitation of cells is one of the most common basic problems in life science research. The established cell detection techniques mainly include fluorescent dye labeling, colorimetric assay, and lateral flow assay, all of which employ antibodies as cell recognition elements. However, the widespread application of the established methods generally dependent on antibodies is limited, because the preparation of antibodies is complicated and time-consuming, and unrecoverable denaturation is prone to occur with antibodies. By contrast, aptamers that are generally selected through the systematic evolution of ligands by exponential enrichment can avoid the disadvantages of antibodies due to their controllable synthesis, thermostability, and long shelf life, etc. Accordingly, aptamers may serve as novel molecular recognition elements like antibodies in combination with various techniques for cell detection. This paper reviews the developed aptamer-based cell detection methods, mainly including aptamer-fluorescent labeling, aptamer-isothermal amplification assay, electrochemical aptamer sensor, aptamer-based lateral flow analysis, and aptamer-colorimetric assay. The principles, advantages, progress of application in cell detection and future development trend of these methods were specially discussed. Overall, different assays are suitable for different detection purposes, and the development of more accurate, economical, efficient, and rapid aptamer-based cell detection methods is always on the road in the future. This review is expected to provide a reference for achieving efficient and accurate detection of cells as well as improving the usefulness of aptamers in the field of analytical applications.









Similar content being viewed by others
Data Availability
All data are available from the authors upon reasonable request.
Abbreviations
- RNA:
-
Ribonucleic acid
- DNA:
-
Deoxyribonucleic acid
- SELEX:
-
Systematic evolution of ligands by exponential enrichment
- WC-SELEX:
-
Whole-cell SELEX
- PCR:
-
Polymerase chain reaction
- LIGS:
-
Ligand-guided selection
- mAb:
-
Monoclonal antibody
- mIgM:
-
Membrane-bound immunoglobulin
- ssDNA:
-
Single-stranded DNA
- HPLC:
-
High-performance liquid chromatography
- PH:
-
Potential of hydrogen
- LAMP:
-
Loop-mediated isothermal amplification
- RCA:
-
Rolling circle amplification
- HCR:
-
Hybridization chain reaction
- L. monocytogenes :
-
Listeria monocytogenes
- PLP:
-
Padlock probe
- DNTPs:
-
Deoxynucleoside triphosphates
- cDNA:
-
Complementary deoxyribonucleic acid
- V. parahaemolyticus :
-
Vibrio Parahaemolyticus
- IC:
-
Initiating chain
- HP:
-
Hairpin probes
- Aptasensors:
-
Aptamer sensors
- E. coli :
-
Escherichia coli
- CFU:
-
Colony forming units
- LFA:
-
Lateral flow assay
- GC:
-
Gas chromatography
- GC-MS:
-
Gas chromatography-mass spectrometry
- LC-MS:
-
Liquid chromatography-mass spectrometry
- V. fischeri :
-
Vibrio fischeri
- ATP:
-
Adenosine triphosphate
- AuNPs:
-
Amphiphilic gold nanoparticles
- GNPs:
-
Gold nanoparticles
- SPR:
-
Surface plasmon resonanceATO
- UV:
-
Ultraviolet
- S. typhimurium :
-
Salmonella typhimurium
- S. aureus :
-
Staphylococcus aureus
- dsDNA:
-
Double-stranded DNA
- SGI:
-
SYBR Green I
References
Zheng LY, Zhang Q, Zhang YT, Qiu LP, Tan WH (2022) Aptamer-based cell recognition and detection. Curr Anal Chem 18(6):612–621
Jin BR, Yang YX, He RY et al (2018) Lateral flow aptamer assay integrated smartphone-based portable device for simultaneous detection of multiple targets using upconversion nanoparticles. Sens Actuat B-Chem 276:48–56
Zhao ZL, Wang H, Zhai WL, Feng XY, Fan X, Chen AL, Wang M (2020) A lateral flow strip based on a truncated aptamer-complementary strand for detection of type-b aflatoxins in nuts and dried figs. Toxins 12(2):136
Huang L, Tian SL, Zhao WH, Liu K, Ma X, Guo JH (2021) Aptamer-based lateral flow assay on-site biosensors. Biosens Bioelectron 186:113279
Stoltenburg R, Reinemann C, Strehlitz B (2007) SELEX-A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 24(4):381–403
Sefah K, Shangguan D, Xiong XL, O’Donoghue MB, Tan WH (2010) Development of DNA aptamers using Cell-SELEX. Nat Protoc 5(6):1169–1185
Batool S, Argyropoulos KV, Azad R, Okeoma P, Zumrut H, Bhandari S, Dekhang R, Mallikaratchy PR (2019) Dimerization of an aptamer generated from ligand-guided selection (LIGS) yields a high affinity scaffold against B-cells. Bba-Gen Subj 1863(1):232–240
Zumrut HE, Ara MN, Maio GE, Van NA, Batool S, Mallikaratchy PR (2016) Ligand-guided selection of aptamers against T-cell receptor-cluster of differentiation 3 (TCR-CD3) expressed on Jurkat.E6 cells. Anal Biochem 512:1–7
Zumrut HE, Ara MN, Fraile M, Maio G, Mallikaratchy P (2016) Ligand-guided selection of Target-Specific Aptamers: a Screening Technology for identifying specific aptamers against cell-surface proteins. Nucleic Acid Ther 26(3):190–198
Chen AL, Yang SM (2015) Replacing antibodies with aptamers in lateral flow immunoassay. Biosens Bioelectron 71:230–242
Wang GQ, Chen ZP, Chen LX (2010) Aptamer-nanoparticle-based optical probes. Prog Chem 22(2–3):489–499
Ozer A, Pagano JM, Lis JT (2014) New Technologies provide Quantum changes in the Scale, Speed, and success of SELEX Methods and Aptamer characterization. Mol Ther-Nucl Acids 3:e183
Sharma TK, Shukla R (2014) Nucleic acid aptamers as an emerging diagnostic tool for animal pathogens. AVS 2(1):50–55
Wang K, Tao ZH, Xu L, Liu YQ (2014) Research and development of functionalized aptamer based biosensor. Chin J Anal Chem 42(2):298–304
Liu JW, Cao ZH, Lu Y (2009) Functional nucleic acid sensors. Chem Rev 109(5):1948–1998
Zhao Q, Geng X, Wang HL (2013) Fluorescent sensing ochratoxin A with single fluorophore-labeled aptamer. Anal Bioanal Chem 405(19):6281–6286
Xu JR, Xiang JH, Chen JL, Wan T, Deng HL, Li DR (2022) High sensitivity detection of tumor cells in biological samples using a multivalent aptamer strand displacement strategy. Analyst 147(4):634–644
Yang L, Meng L, Zhang XB, Chen Y, Zhu GZ, Liu HP, Xiong XL, Sefah K, Tan WH (2011) Engineering polymeric aptamers for selective cytotoxicity. J Am Chem Soc 133(34):13380–13386
Savla R, Taratula O, Garbuzenko O, Minko T (2011) Tumor targeted quantum dot-mucin 1 aptamer-doxorubicin conjugate for imaging and treatment of cancer. J Control Release 153(1):16–22
Zhang J, Jia X, Lv XJ, Deng YL, Xie HY (2010) Fluorescent quantum dot-labeled aptamer bioprobes specifically targeting mouse liver cancer cells. Talanta 81(1–2):505–509
Hu ZX, Tan JT, Lai ZQ et al (2017) Aptamer combined with fluorescent silica nanoparticles for detection of hepatoma cells. Nanoscale Res Lett 12(1):1–8
Feng J, Shan GM, Maquieira A, Koivunen ME, Guo B, Hammock BD, Kennedy IM (2003) Functionalized europium oxide nanoparticles used as a fluorescent label in an immunoassay for atrazine. Anal Chem 75(19):5282–5286
Zhao Y, Zuo XL, Li Q et al (2021) Nucleic acids analysis. Sci China Chem 64(2):171–203
Yang H, Xu DK (2022) Highly-sensitive and simple fluorescent aptasensor for 17 b-estradiol detection coupled with HCR-HRP structure. Talanta 240:123094
Zhao YX, Chen F, Li Q, Wang LH, Fan CH (2015) Isothermal amplification of nucleic acids. Chem Rev 115(22):12491–12545
Wilson DS, Szostak JW (1999) In vitro selection of functional nucleic acids. Annu Rev Biochem 68:611–647
Tan WH, Donovan MJ, Jiang JH (2013) Aptamers from cell-based selection for Bioanalytical Applications. Chem Rev 113(4):2842–2862
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. NAR 28(12):e63
Nagamine K, Hase T, Notomi T (2002) Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probe 16(3):223–229
Feng JL, Dai ZY, Tian XL, Jiang XN (2018) Detection of Listeria monocytogenes based on combined aptamers magnetic capture and loop-mediated isothermal amplification. Food Control 85:443–452
Schweitzer B, Wiltshire S, Lambert J, O’Malley S, Kukanskis K, Zhu ZR, Kingsmore SF, Lizardi PM, Ward DC (2000) Immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. P Natl Acad Sci USA 97(18):10113–10119
Kingsmore SF, Patel DD (2003) Multiplexed protein profiling on antibody-based microarrays by rolling circle amplification. Curr Opin Biotech 14(1):74–81
Qi XQ, Bakht S, Devos KM, Gale MD, Osbourn A (2001) L-RCA (ligation-rolling circle amplification): a general method for genotyping of single nucleotide polymorphisms (SNPs). NAR 29(22):e116
Deng RJ, Tang LH, Tian QQ, Wang Y, Lin L, Li JH (2014) Toehold-initiated Rolling Circle amplification for visualizing individual MicroRNAs in situ in single cells. Angew Chem Int Edit 53(9):2389–2393
Asadi R, Mollasalehi H (2021) The mechanism and improvements to the isothermal amplification of nucleic acids, at a glance. Anal Biochem 631:114260
Xu XY, Su Y, Zhang YZ, Wang XX, Tian HT, Ma X, Chu HS, Xu WT (2021) Novel rolling circle amplification biosensors for food-borne microorganism detection. TrAC-Trend Anal Chem 141:116293
Dirks RM, Pierce NA (2004) Triggered amplification by hybridization chain reaction. P Natl Acad Sci USA 101(43):15275–15278
Zhou GB, Lin MH, Song P, Chen XQ, Chao J, Wang LH, Huang Q, Huang W, Fan CH, Zuo XL (2014) Multivalent capture and detection of Cancer cells with DNA nanostructured biosensors and multibranched hybridization chain reaction amplification. Anal Chem 86(15):7843–7848
Zhang C, Belwal T, Luo ZS, Su B, Lin XY (2021) Application of Nanomaterials in Isothermal Nucleic Acid amplification. Small 18(6):2102711
Li L, Jiang HS, Meng XX, Wen XH, Guo QP, Li ZH, Wang J, Ren YZ, Wang KM (2020) Highly sensitive detection of cancer cells via split aptamer mediated proximity-induced hybridization chain reaction. Talanta 223:121724
Liu YX, Chen P, Yuan S, Sun B, Sun R, Meng XH (2020) A novel method for sensitive detection of Escherichia coli O157:H7 based on an aptamer and hybridization chain reaction. Anal Methods 12(29):3734–3740
Yuan YL, Wei SQ, Liu GP, Xie SB, Chai YQ, Yuan R (2014) Ultrasensitive electrochemiluminescent aptasensor for ochratoxin A detection with the loop-mediated isothermal amplification. Anal Chim Acta 811:70–75
Xu LL, Duan JX, Chen JM, Ding SJ, Cheng W (2020) Recent advances in rolling circle amplification-based biosensing strategies-A review. Anal Chim Acta 1148:238187
Bi S, Yue SZ, Zhang SS (2017) Hybridization chain reaction: a versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem Soc Rev 46(14):4281–4298
Abu H, Hossain M, Marlinda A, Al Mamun M, Simarani K, Johan MR (2021) Nanomaterials based electrochemical nucleic acid biosensors for environmental monitoring: a review. Appl Surf Sci Adv 4:100064
Barthelmebs L, Hayat A, Limiadi AW, Marty JL, Noguer T (2011) Electrochemical DNA aptamer-based biosensor for OTA detection, using superparamagnetic nanoparticles. Sens Actuat B-Chem 156(2):932–937
Du Y, Dong SJ (2017) Nucleic acid biosensors: recent advances and perspectives. Ana Chem 89(1):189–215
Ziolkowski R, Jarczewska M, Gorski L, Malinowska E (2021) From small molecules toward whole cells detection: application of Electrochemical Aptasensors in Modern Medical Diagnostics. Sensors 21(3):724
Majdinasab M, Yaqub M, Rahim A, Catanante G, Hayat A, Marty JL (2017) An overview on recent progress in electrochemical biosensors for antimicrobial drug residues in animal-derived food. Sensors 17(9):1947
Liu MY, Yue FL, Kong QQ, Liu ZL, Guo YM, Sun X (2022) Aptamers against pathogenic Bacteria: selection strategies and Apta-assay/Aptasensor application for Food Safety. J Agr Food Chem 70(18):5477–5498
Ranjbar S, Shahrokhian S, Nurmohammadi F (2018) Nanoporous gold as a suitable substrate for preparation of a new sensitive electrochemical aptasensor for detection of Salmonella typhimurium. Sens Actuat B-Chem 255:1536–1544
Hasan MR, Pulingam T, Appaturi JN, Zifruddin AN, Teh SJ, Lim TW, Ibrahim F, Leo BF, Thong KL (2018) Carbon nanotube-based aptasensor for sensitive electrochemical detection of whole-cell Salmonella. Anal Biochem 554:34–43
Burrs SL, Bhargava M, Sidhu R, Kiernan-Lewis J, Gomes C, Claussen JC, McLamore ES (2016) A paper based graphene-nanocauliflower hybrid composite for point of care biosensing. Biosens Bioelectron 85:479–487
Reich P, Stoltenburg R, Strehlitz B, Frense D, Beckmann D (2017) Development of an impedimetric aptasensor for the detection of Staphylococcus aureus. Int J Mol Sci 18(11):2484
Tian L, Qi JX, Qian K, Oderinde O, Liu QY, Yao C, Song W, Wang YH (2018) Copper (II) oxide nanozyme based electrochemical cytosensor for high sensitive detection of circulating tumor cells in breast cancer. J Electroanal Chem 812:1–9
Yang YB, Yang XD, Yang YJ, Yuan Q (2018) Aptamer-functionalized carbon nanomaterials electrochemical sensors for detecting cancer relevant biomolecules. Carbon 129:380–395
Nuntawong P, Putalun W, Tanaka H, Morimoto S, Sakamoto S (2022) Lateral flow immunoassay for small-molecules detection in phytoproducts: a review. J Nat Med-Tokyo 76(3):521–545
Wang T, Chen LM, Chikkanna A, Chen SX, Brusius I, Sbuh N, Veedu RN (2021) Development of nucleic acid aptamer-based lateral flow assays: a robust platform for cost-effective point-of-care diagnosis. Theranostics 11(11):5174–5196
Reid R, Chatterjee B, Das SJ, Ghosh S, Sharma TK (2020) Application of aptamers as molecular recognition elements in lateral flow assays. Anal Biochem 593:113574
Bahadır EB, Sezgintürk MK (2016) Lateral flow assays: principles, designs and labels. TrAC 82:286–306
Posthuma-Trumpie GA, Korf J, van Amerongen A (2009) Lateral flow (immuno)assay: its strengths, weaknesses, opportunities and threats. A literature survey. Anal Bioanal Chem 393(2):569–582
Giblin PA, Lemieux RM (2006) LFA-1 as a key regulator of immune function: approaches toward the development of LFA-1-based therapeutics. Curr Pharm Design 12(22):2771–2795
Sajid M, Kawde AN, Daud M (2015) Designs, formats and applications of lateral flow assay: a literature review. J Saudi Chem Soc 19(6):689–705
Jauset-Rubio M, El-Shahawi MS, Bashammakh AS, Alyoubi AO, O’Sullivan CK (2017) Advances in aptamers-based lateral flow assays. TrAC 97:385–398
Dalirirad S, Steckl AJ (2020) Lateral flow assay using aptamer-based sensing for on-site detection of dopamine in urine. Anal Biochem 596:113637
Shin WR, Sekhon SS, Rhee SK, Ko JH, Ahn JY, Min J, Kim YH (2018) Aptamer-based paper Strip Sensor for detecting Vibrio fischeri. Acs Comb Sci 20(5):261–268
Liu GD, Mao X, Phillips JA, Xu H, Tan WH, Zeng LW (2009) Aptamer-nanoparticle strip biosensor for sensitive detection of cancer cells. Anal Chem 81(24):10013–10018
Nam JM, Park SJ, Mirkin CA (2002) Bio-barcodes based on oligonucleotide-modified nanoparticles. J Am Chem Soc 124(15):3820–3821
Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA (1997) Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 277(5329):1078–1081
Pavlov V, Xiao Y, Shlyahovsky B, Willner I (2004) Aptamer-functionalized au nanoparticles for the amplified optical detection of thrombin. J Am Chem Soc 126(38):11768–11769
Liu JW, Lu Y (2003) A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J Am Chem Soc 125(22):6642–6643
Zhao WA, Chiuman W, Lam J, McManus SA, Chen W, Cui YG, Pelton R, Brook MA, Li YF (2008) DNA aptamer folding on gold nanoparticles: from colloid chemistry to biosensors. J Am Chem Soc 130(11):3610–3618
Medley CD, Smith JE, Tang Z, Wu Y, Bamrungsap S, Tan WH (2008) Gold nanoparticle-based colorimetric assay for the direct detection of cancerous cells. Anal Chem 80(4):1067–1072
Wu SJ, Wang YQ, Duan N, Ma HL, Wang ZP (2015) Colorimetric Aptasensor based on enzyme for the detection of Vibrio parahemolyticus. J Agr Food Chem 63(35):7849–7854
Ma XY, Song LJ, Zhou NX, Xia Y, Wang ZP (2017) A novel aptasensor for the colorimetric detection of S. typhimurium based on gold nanoparticles. Int J Food Microbiol 245:1–5
Yu TX, Xu H, Zhao Y, Han YJ, Zhang Y, Zhang JR, Xu CH, Wang WJ, Guo Q, Ge J (2020) Aptamer based high throughput colorimetric biosensor for detection of staphylococcus aureus. Sci Rep-UK 10(1):1–6
Xie MJ, Zhao FG, Zhang YP, Xiong Y, Han SY (2022) Recent advances in aptamer-based optical and electrochemical biosensors for detection of pesticides and veterinary drugs. Food Control 131:108399
Funding
This work was supported by Shandong Provincial Natural Science Foundation, China (ZR2020MD081); the National Scientific Foundation of China (No. 31600309, 41476086); HIT Scientific Research Innovation Fund (No. 2022KYCXJJ07); and HIT Environment and Ecology Innovation Special Funds (No. HSCJ201622).
Author information
Authors and Affiliations
Contributions
Conceived and designed the paper: Fuguo Liu, Chunyun Zhang and Guofu Chen; Analysis and arrangement of documents: Yu Duan, Jinju Ma, Yuanyuan Wang and Wenrong Chen; Wrote the paper: Wenrong Chen.
Corresponding authors
Ethics declarations
Competing Interests
We seriously declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work. There is also no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in or the review of the manuscript entitled “A review of advances in aptamer-based cell detection technology”.
Statements & Declarations
All the authors would like to seriously state that: (a) the paper is not currently being considered for publication elsewhere; (b) all authors have been personally and actively involved in substantive work leading to the report, and will hold themselves jointly and individually responsible for its content; and (c) all relevant ethical safeguards have been met in this study.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Consent to Publish
All the authors consent to publish all the data included in the paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, W., Liu, F., Zhang, C. et al. A review of advances in aptamer-based cell detection technology. Mol Biol Rep 50, 5425–5438 (2023). https://doi.org/10.1007/s11033-023-08410-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08410-8