Abstract
Background
Alzheimer’s disease (AD) is an incurable and debilitating neurodegenerative disease that results in the progressive degeneration and death of nerve cells. Mutations in the APP gene, which encodes an amyloid precursor protein, is the strongest genetic risk factor for sporadic AD.
Methods and Results
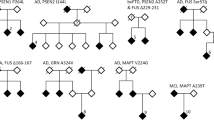
We studied the APP gene (NM_000484.3: c.2045A > T; p.E682V) variants carried by members of a family suffering from AD using whole-exome sequencing and Sanger sequencing.
Conclusion
In this study, we identified a new variant of the APP gene (NM_000484.3: c.2045A > T; p.E682V) in members of a family with AD. This provides potential targets for subsequent studies and information that can be used in genetic counselling.


Similar content being viewed by others
References
Wlodarek D (2019) Role of ketogenic diets in neurodegenerative diseases (Alzheimer’s disease and Parkinson’s disease). Nutrients 11(1):169
Ghani M et al (2015) Mutation analysis of patients with neurodegenerative disorders using NeuroX array. Neurobiol Aging 36(1):5459–5514
Masters CL et al (2015) Alzheimer’s disease. Nat Rev Dis Primers 1:15056
Bai B et al (2021) Proteomic landscape of Alzheimer’s disease: novel insights into pathogenesis and biomarker discovery. Mol Neurodegener 16(1):55
Moussa-Pacha NM et al (2020) BACE1 inhibitors: current status and future directions in treating Alzheimer’s disease. Med Res Rev 40(1):339–384
Serrano-Pozo A, Das S, Hyman BT (2021) APOE and Alzheimer’s disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol 20(1):68–80
Zhou L et al (2011) Amyloid precursor protein mutation E682K at the alternative beta-secretase cleavage beta’-site increases Abeta generation. EMBO Mol Med 3(5):291–302
Yao Y et al (2021) A delta-secretase-truncated APP fragment activates CEBPB, mediating Alzheimer’s disease pathologies. Brain 144(6):1833–1852
Hung C, Livesey FJ (2021) Endolysosome and autophagy dysfunction in Alzheimer disease. Autophagy 17(11):3882–3883
Graff-Radford J et al (2021) New insights into atypical Alzheimer’s disease in the era of biomarkers. Lancet Neurol 20(3):222–234
Chhatwal JP et al (2022) Variant-dependent heterogeneity in amyloid beta burden in autosomal dominant Alzheimer’s disease: cross-sectional and longitudinal analyses of an observational study. Lancet Neurol 21(2):140–152
Scheltens P et al (2021) Alzheimer’s disease. Lancet 397(10284):1577–1590
Van Bulck M et al (2019) Novel approaches for the treatment of Alzheimer’s and Parkinson’s disease. Int J Mol Sci 20(3):719
Manczak M et al (2018) Hippocampal mutant APP and amyloid beta-induced cognitive decline, dendritic spine loss, defective autophagy, mitophagy and mitochondrial abnormalities in a mouse model of Alzheimer’s disease. Hum Mol Genet 27(8):1332–1342
Hur JY (2022) Gamma-secretase in Alzheimer’s disease. Exp Mol Med 54(4):433–446
Jorda-Siquier T et al (2022) APP accumulates with presynaptic proteins around amyloid plaques: a role for presynaptic mechanisms in Alzheimer’s disease? Alzheimers Dement 18(11):2099–2116
Jia L et al (2020) PSEN1, PSEN2, and APP mutations in 404 Chinese pedigrees with familial Alzheimer’s disease. Alzheimers Dement 16(1):178–191
Wang CY et al (2020) A novel nonsense mutation of ABCA8 in a Han-Chinese family With ASCVD leads to the reduction of HDL-c levels. Front Genet 11:755
Galvao F Jr et al (2019) The amyloid precursor protein (APP) processing as a biological link between Alzheimer’s disease and cancer. Ageing Res Rev 49:83–91
Nikolac Perkovic M, Pivac N (2019) Genetic markers of Alzheimer’s disease. Adv Exp Med Biol 1192:27–52
Cacace R, Sleegers K, Van Broeckhoven C (2016) Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimers Dement 12(6):733–748
Brouwers N, Sleegers K, Van Broeckhoven C (2008) Molecular genetics of Alzheimer’s disease: an update. Ann Med 40(8):562–583
Liu YC et al (2021) Melatonin Induction of APP intracellular domain 50 SUMOylation alleviates AD through enhanced transcriptional activation and Abeta degradation. Mol Ther 29(1):376–395
Acknowledgements
We thank all authors for their help in the completion of this paper and the medical staff of the department for their good service to the participants. The authors are thankful to the anonymous reviews for the constructive criticism of this article.
Funding
This study was supported by the National Natural Science Foundation of China (82000427).
Author information
Authors and Affiliations
Contributions
LX and TT enrolled the samples and clinical data. WZ isolated the gDNA and performed PCR. YZ in performed genetic analysis. WC wrote the manuscript. FL supported the study. All authors reviewed the manuscript. Each author participated sufficiently and consent to publication.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Ethical approval
This study was approved and supervised by the ethics committee of the Yangzhou Oriental Hospital. All individuals who participated in the study were given written informed consent forms. Considering the serious symptoms of the proband, we invited the son as guardian to sign the informed consent form. We also obtained brain scans with permission to publish from the legal representative of the proband.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhaoxia, W., Chenyu, W., ZhuangZhuang, Y. et al. Whole-exome sequencing detected a novel APP variant in a Han-Chinese family with Alzheimer’s disease. Mol Biol Rep 50, 5267–5271 (2023). https://doi.org/10.1007/s11033-023-08400-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08400-w