Abstract
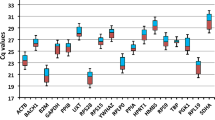
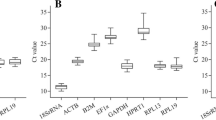
Coturniculture has been standing out as an industrial poultry activity in several countries around the world because of the several adaptive advantages of quails. Research that considers the analysis of gene expression can enhance this activity. This study aimed to analyze the stability of reference genes (RGs) in different tissues of quails (both males and females) for the recommendation of use in gene expression studies by the quantitative reverse transcription-polymerase chain reaction (RT-qPCR). The expression stability of ten RGs (ACTA1, ACTB, B2M, GAPDH, HMBS, SDHA, HPRT1, MRPS27, MRPS30, and RPL5) was analyzed in four tissues (breast muscle, abdominal fat, liver, and intestine), and assessed using the statistical tools geNorm, NormFinder, comparative ΔCq method, and BestKeeper. The HPRT1 gene was the most stable in all quail tissues tested, followed by MRPS27 and MRPS30 in breast muscle, B2M and RPL5 in abdominal fat, HMBS and B2M in the liver, and RPL5 and HMBS in the intestine. These results may help studies using RT-qPCR assays to assess quail tissues from both sexes because they provide data on the most stable genes, which should be tested as candidate RGs for other experimental conditions.


Similar content being viewed by others
References
Barrete TLS, Quirino BJS, Brito CD, Umigi RT, Araújo MS, Coimbra JRS, Rojas GEE, Freitas FFJ, Reis SR (2007) Metabolizable energy levels for Japanese quails in the initial laying phase. Rev Bras Zoot 36:79–85. https://doi.org/10.1590/S1516-35982007000100010
Lukanov H (2019) Domestic quail (Coturnix japonica domestica), is there such farm animal? W Poul Sci J 75:547–558. https://doi.org/10.1017/S0043933919000631
López-Salazar SE, Flota-Bañuelos C, Fraire-Cordero S (2020) Agroecological production of codorniz (Coturnix coturnix japonica) as strategy for food security in Campeche, Mexico. Agroproductividad 13:3–7. https://doi.org/10.32854/agrop.vi0.1513
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Vandesompele J (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. C Chem 55:611–622. https://doi.org/10.1373/clinchem.2008.112797
Shukla P, Reddy RA, Ponnuvel KM, Rohela GK, Shabnam AA, Ghosh MK, Mishra RK (2019) Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Mulberry (Morus alba L.) under different abiotic stresses. Mol Biol Rep 46:1809–1817. https://doi.org/10.1007/s11033-019-04631-y
Wang B, Lirong WANG, Duan H, Chong P, Su S, Shan L, Li Y (2020) Selection and validation of reference genes for quantitative real-time PCR analysis of Nitraria tangutorum. https://doi.org/10.21203/rs.2.21923/v1
Pfaffl MW, Tichopad A, Prgomet C (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515. https://doi.org/10.1023/B:BILE.0000019559.84305.47
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalization; strategies and considerations. G Immun 6:279–284. https://doi.org/10.1038/sj.gene.6364190
Sanders R, Mason DJ, Foy CA, Huggett JF (2014) Considerations for accurate gene expression measurement by reverse transcription quantitative PCR when analysing clinical samples. Anal Bioanal Chem 406:6471–6483. https://doi.org/10.1007/s00216-014-7857-x
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCRÐA perspective. J Mol Endoc 34:597–601. https://doi.org/10.1677/jme.1.01755
Tang B, Dai W, Zhang C (2019) Selection of reference genes for quantitative real-time polymerase chain reaction normalization in Bradysia odoriphaga (Diptera: Sciaridae). Entom Sci 22:422–436. https://doi.org/10.1111/ens.12383
Lee PD, Sladek R, Greenwood CM, Hudson TJ (2002) Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genon Res 12:292–297. https://doi.org/10.1101/gr.217802
Nascimento CS, Barbosa LT, Brito C, Fernandes RP, Mann RS, Pinto APG, Oliveira HC, Dodson MV, Guimarães SEF, Duarte MS (2015) Identification of suitable reference genes for real time quantitative polymerase chain reaction assays on pectoralis major muscle in chicken (Gallus gallus). PLoS One 10:e0127935. https://doi.org/10.1371/journal.pone.0127935
Jemiolo B, Trappe S (2004) Single muscle fiber gene expression in human skeletal muscle: validation of internal control with exercise. Bioch Biophys Res Commun 320:1043–1050. https://doi.org/10.1016/j.bbrc.2004.05.223
Axtner J, Sommer S (2009) Validation of internal reference genes for quantitative real-time PCR in a non-model organism, the yellow-necked mouse, Apodemus flavicollis. BMC Res Note 2:1–7. https://doi.org/10.1186/1756-0500-2-264
Weyrich A, Axtner J, Sommer S (2010) Selection and validation of reference genes for realtime RT-PCR studies in the non-model species Delomys sublineatus, an endemic Brazilian rodent. Bioch Biophys Res Commun 392:145–149. https://doi.org/10.1016/j.bbrc.2009.12.173
Zhang Y, Han X, Chen S, Zheng L, He X, Liu M, Zhuo R (2017) Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Salix matsudana under different abiotic stresses. Sci Report 7:1–11. https://doi.org/10.1038/srep40290
Li Z, Lu H, He Z, Wang C, Wang Y, Ji X (2019) Selection of appropriate reference genes for quantitative real-time reverse transcription PCR in Betula platyphylla under salt and osmotic stress conditions. PLoS One 14:e0225926. https://doi.org/10.1371/journal.pone.0225926
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Gen Biol 7:1–12. https://doi.org/10.1186/gb-2002-3-7-research0034
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Can Res 64:5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496
Silver N, Best S, Jiang J (2006) Selection of housekeeping genes for genes expression studies in human reticulocytes using real-time PCR. BMC Mol Bio 7:1–7. https://doi.org/10.1186/1471-2199-7-33
Lisowski P, Pierzchała M, Gościk J, Pareek CS, Zwierzchowski L (2008) Evaluation of reference genes for studies of gene expression in the bovine liver, kidney, pituitary, and thyroid. J Appl Gen 49:367–372 https://doi.org/10.100/BF03195635
Bonnet M, Bernard L, Bes S, Leroux C (2013) Selection of reference genes for quantitative real-time PCR normalisation in adipose tissue, muscle, liver and mammary gland from ruminants. Animal 7:1344–1353. https://doi.org/10.1017/S1751731113000475
Cheng L, Yu J, Hu X, Xiang M, Xia Y, Tao B, Chen H (2020) Identification of reliable reference genes for expression studies in maternal reproductive tissues and fetal tissues of pregnant cows. R Dom Anim. https://doi.org/10.1111/rda.13808
Martínez-Giner M, Noguera JL, Balcells I, Fernández-Rodríguez A, Pena RN (2013) Selection of internal control genes for real-time quantitative PCR in ovary and uterus of sows across pregnancy. PLoS One 8:e66023. https://doi.org/10.1371/journal.pone.0066023
Zang R, Bai J, Xu H, Zhang L, Yang J, Yang L, Lu J, Wu J (2011) Selection of suitable reference genes for real-time quantitative PCR studies in Lanzhou fat-tailed sheep (Ovis ares). Asi J A Vet Advan Y 6:789–804. https://doi.org/10.3923/ajava.2011.789.804
Najafpanah MJ, Sadeghi M, Bakhtiarizadeh MR (2013) Reference genes selection for quantitative real-time PCR using RankAggreg method in different tissues of Capra hircus. PLoS One 8:e83041. https://doi.org/10.1371/journal.pone.0083041
Katarzyńska-Banasik D, Grzesiak M, Sechman A (2017) Selection of reference genes for quantitative real-time PCR analysis in chicken ovary following silver nanoparticle treatment. Environ Toxicol Pharmacol 56:186–190. https://doi.org/10.1016/j.etap.2017.09.011
Yang CG, Wang XL, Tian J, Liu W, Wu F, Jiang M, Wen H (2013) Evaluation of reference genes for quantitative real-time RT-PCR analysis of gene expression in Nile tilapia (Oreochromis niloticus). Gene 527:183–192. https://doi.org/10.1016/j.gene.2013.06.013
Ahn K, Bae JH, Nam KH, Lee CE, Park KD, Lee HK, Cho BW, Kim HS (2011) Identification of reference genes for normalization of gene expression in thoroughbred and Jeju native horse (Jeju pony) tissues. G Gen 33:245–250. https://doi.org/10.1007/s13258-010-0114-6
Ashish S, Bhure SK, Harikrishna P, Ramteke SS, Kutty VM, Shruthi N, Mihir S (2017) Identification and evaluation of reference genes for accurate gene expression normalization of fresh and frozen-thawed spermatozoa of water buffalo (Bubalus bubalis). Theriogenology 92:6–13. https://doi.org/10.1016/j.theriogenology.2017.01.006
Kaur R, Sodhi M, Sharma A, Sharma VL, Verma P, Swami SK, Mukesh M (2018) Selection of suitable reference genes for normalization of quantitative RT-PCR (RT-qPCR) expression data across twelve tissues of riverine buffaloes (Bubalus bubalis). PLoS One 13:e0191558. https://doi.org/10.1371/journal.pone.0191558
Wu Y, Zhang Y, Hou Z, Fan G, Pi J, Sun S, Hu G (2018) Population genomic data reveal genes related to important traits of quail. GigaScience 7:1–16. https://doi.org/10.1093/gigascience/giy049
Carvalho A, Couroussé N, Crochet S, Coustham V (2019) Identification of reference genes for quantitative gene expression studies in three tissues of Japanese Quail. Genes 10:1–12. https://doi.org/10.3390/genes10030197
Olias P, Adam I, Meyer A, Scharff C, Gruber AD (2014) Reference genes for quantitative gene expression studies in multiple avian species. PLoS One 9:e99678. https://doi.org/10.1371/journal.pone.0099678
Oliveira H, Garcia AA, Gromboni JG, Farias Filho R, Nascimento C, Wenceslau AA (2017) Influence of heat stress, sex and genetic groups on reference genes stability in muscle tissue of chicken. PLoS One 12:e0176402. https://doi.org/10.1371/journal.pone.0176402
Hassanpour H, Aghajani Z, Bahadoran S, Farhadi N, Nazari H, Kaewduangta W (2019) Identification of reliable reference genes for quantitative real-time PCR in ovary and uterus of laying hens under heat stress. Stress 22:387–394. https://doi.org/10.1080/10253890.2019.1574294
Pfaffl MWA (2001) A new mathematical model for relative quantification in real-time RT- PCR. Nucl A Res 29:e45. https://doi.org/10.1093/nar/29.9.e45
Livak JK, Schmittgen DT (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Chapman JR, Helin AS, Wille M, Atterby C, Järhul JD, Fridlund JS, Waldenström J (2016) A panel of stably expressed reference genes for real-time qPCR gene expression studies of mallards (Anas platyrhynchos). PLoS One 11:e0149454. https://doi.org/10.1371/journal.pone.0149454
Nygard AB, Jørgensen CB, Cirera S, Fredholm M (2007) Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Bio 8:1–6. https://doi.org/10.1186/1471-2199-8-67
Hildyard JC, Wells DJ (2014) Identification and validation of quantitative PCR reference genes suitable for normalizing expression in normal and dystrophic cell culture models of myogenesis. PLoS One 11:e0149454. https://doi.org/10.1371/currents.md.faafdde4bea8df4aa7d06cd5553119a6
Feng X, Xiong Y, Qian H, Lei M, Xu D, Ren Z (2010) Selection of reference genes for gene expression studies in porcine skeletal muscle using SYBR green qPCR. J Biotec 150:288–293. https://doi.org/10.1016/j.jbiotec.2010.09.949
Chapman JR, Waldenström J (2015) With reference to reference genes: a systematic review of endogenous controls in gene expression studies. PLoS One 10:e0141853. https://doi.org/10.1371/journal.pone.0141853
Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR (2001) β-Actin an unsuitable internal control for RT-PCR. Mol Cell Probes 15:307–311. https://doi.org/10.1006/mcpr.2001.0376
Li X, Huang K, Chen F, Li W, Sun S, Shi XE, Yang G (2016) Verification of suitable and reliable reference genes for quantitative real-time PCR during adipogenic differentiation in porcine intramuscular stromal-vascular cells. Animal 10:947–952. https://doi.org/10.1017/S1751731115002748
Cedraz de Oliveira H, Pinto Garcia AA, Gonzaga Gromboni JG, Vasconcelos Farias Filho R, Souza do Nascimento C, Arias Wenceslau A (2017) Influence of heat stress, sex and genetic groups on reference genes stability in muscle tissue of chicken. PLoS One 12:e0176402. https://doi.org/10.1371/journal.pone.0176402
Funding
We thank the Fundação CAPES (Finance code 001) and Fundação de Amparo à Pesquisa do Estado do Piauí (FAPEPI) for the financial support.
Author information
Authors and Affiliations
Contributions
FCBS, CSN, MSM, LTB, GFVB, KRSS: conceived and the study; FCBS, CSN, MSM, LTB: conducted the experiments; FCBS, CSN, RSA, GFVB, KRSS: analyzed the data; FCBS, RSA, KRSS: wrote the manuscript. All authors read and approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures applied in this study were approved by the Animal Ethics and Protection Committee of the Federal University of Sergipe (Universidade Federal de Sergipe), Sergipe, Brazil.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
de Sousa, F.C.B., do Nascimento, C.S., Macário, M.d.S. et al. Selection of reference genes for quantitative real-time PCR normalization in European quail tissues. Mol Biol Rep 48, 67–76 (2021). https://doi.org/10.1007/s11033-020-06134-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-06134-7