Abstract
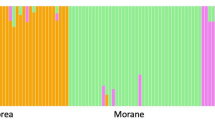
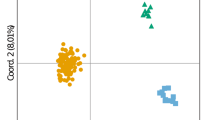
Cetaceans are large mammals widely distributed on Earth. The fin whale, Balaenoptera physalus, is the second largest living animal. In the 20th century, commercial whaling reduced its global population by 70%, and in the Mediterranean Sea not only was their overall population depleted but the migration between the Mediterranean Sea and the Atlantic Ocean was reduced. Previous genetic studies identified isolation between these two regions, with a limited gene-flow between these adjacent populations based on nuclear and mitochondrial markers. However, only limited information exists for the Mediterranean population as genetic diversity and abundance trends are still unknown. In this study, 39 highly polymorphic microsatellite markers were tested, including 25 markers developed de novo together with 14 markers previously published. An average allelic diversity of 8.3 alleles per locus was reported, ranging from 3 to 15 alleles per locus, for B. physalus. Expected heterozygosity was variable among loci and ranged from 0.34 to 0.91. Only two markers in the new set were significantly deviant from the Hardy Weinberg equilibrium. Cross-species amplification was tested in four other cetacean species. A total of 27 markers were successfully amplified in the four species (Balaenoptera acutorostrata, Megaptera novaeangliae, Physeter macrocephalus and Globicephala melas). A multivariate analysis on the multilocus genotypes successfully discriminated the five species. This new set of microsatellite markers will not only provide a useful tool to identify and understand the genetic diversity and the evolution of the B. physalus population, but it will also be relevant for other cetacean species, and will allow further parentage analyses. Eventually, this new set of microsatellite markers will provide critical data that will shed light on important biological data within a conservation perspective.

Similar content being viewed by others
References
Harwood J (2001) Marine mammals and their environment in the twenty-first century. J Mammal 82:630–640
Brownell RL, Yablokov AV (2009) Whaling, illegal and pirate. In: Encyclopedia of marine mammals, 2nd edn. Elsevier, Amsterdam, pp 1235–1239
Cooke JG (2018) Balaenoptera physalus. IUCN Red List Threat Species 2018 eT2478A50349982. https://doi.org/10.2305/IUCN.UK.2018-2.RLTS.T2478A50349982.en
Sanpera C, Aguilar A (1992) Modern whaling off the Iberian Peninsula during the 20th century. Rep Int Whal Comm 42:723–730
Forcada J, Aguilar A, Hammond PS et al (1996) Distribution and abundance of fin whales (Balaenoptera physalus) in the western Mediterranean sea during the summer. J Zool 238:23–34. https://doi.org/10.1111/j.1469-7998.1996.tb05377.x
Notarbartolo-di-Sciara G, Zanardelli M, Jahoda M et al (2003) The fin whale Balaenoptera physalus (L. 1758) in the Mediterranean Sea. Mamm Rev 33:105–150
Bérubé M, Aguilar A, Dendanto D et al (1998) Population genetic structure of North Atlantic, Mediterranean Sea and Sea of Cortez fin whales, Balaenoptera physalus (Linnaeus 1758): analysis of mitochondrial and nuclear loci. Mol Ecol 7:585–599. https://doi.org/10.1046/j.1365-294x.1998.00359.x
Weber JL, May PE (1989) Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet 44:388–396
Lamb CT, Ford AT, Proctor MF et al (2019) Genetic tagging in the Anthropocene: scaling ecology from alleles to ecosystems. Ecol Appl 29:e01876. https://doi.org/10.1002/eap.1876
Clisson I, Lathuilliere M, Crouau-Roy B (2000) Conservation and evolution of microsatellite loci in primate taxa. Am J Primatol 50:205–214. https://doi.org/10.1002/(SICI)1098-2345(200003)50:3%3c205:AID-AJP3%3e3.0.CO;2-Y
Hedgecock D, Li G, Hubert S et al (2004) Widespread null alleles and poor cross-species amplification of microsatellite DNA loci cloned from the pacific oyster, Crassostrea gigas. J Shellfish Res 23:379–385
Crouau-Roy B, Service S, Slatkin M, Freimer N (1996) A fine-scale comparison of the human and chimpanzee genomes: linkage, linkage disequilibrium and sequence analysis. Hum Mol Genet 5:1131–1137. https://doi.org/10.1093/hmg/5.8.1131
Valsecchi E, Amos W (1996) Microsatellite markers for the study of cetacean populations. Mol Ecol 5:151–156. https://doi.org/10.1111/j.1365-294X.1996.tb00301.x
Palsbøll PJ, Bérubé M, Larsen AH, Jørgensen H (1997) Primers for the amplification of tri- and tetramer microsatellite loci in baleen whales. Mol Ecol 6:893–895. https://doi.org/10.1111/j.1365-294X.1997.tb00146.x
Bérubé M, Jørgensen H, McEwing R, Palsbøll PJ (2000) Polymorphic di-nucleotide microsatellite loci isolated from the humpback whale, Megaptera novaeangliae. Mol Ecol 9:2181–2183. https://doi.org/10.1046/j.1365-294X.2000.105315.x
Bérubé M, Rew MB, Skaug H et al (2005) Polymorphic microsatellite loci isolated from humpback whale, Megaptera novaeangliae and fin whale, Balaenoptera physalus. Conserv Genet 6:631–636. https://doi.org/10.1007/s10592-005-9017-5
Meglécz E, Pech N, Gilles A et al (2014) QDD version 3.1: a user-friendly computer program for microsatellite selection and primer design revisited: experimental validation of variables determining genotyping success rate. Mol Ecol Resour 14:1302–1313. https://doi.org/10.1111/1755-0998.12271
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214. https://doi.org/10.1089/10665270050081478
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Peakall R, Smouse PE (2012) GenALEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Belkhir K, Borsa P, Chikhi L, et al (2004) GENETIX: Windows TM software for population genetics. Lab. Génome, Popul. Interact. CNRS Umr 5000, Univ. Montpellier II, Montpellier (France)
Paetkau D, Strobeck C (1995) The molecular basis and evolutionary history of a microsatellite null allele in bears. Mol Ecol 4:519–520. https://doi.org/10.1111/j.1365-294X.1995.tb00248.x
Hayden MJ, Nguyen TM, Waterman A et al (2008) Application of multiplex-ready PCR for fluorescence-based SSR genotyping in barley and wheat. Mol Breed 21:271–281. https://doi.org/10.1007/s11032-007-9127-5
Edwards MC, Gibbs RA (1994) Multiplex PCR: advantages, development, and applications. Genome Res 3:S65–S75. https://doi.org/10.1101/gr.3.4.S65
Bourret V, Macé M, Bonhomme M, Crouau-Roy B (2008) Microsatellites in cetaceans: an overview. Open Mar Biol J 2:38–42. https://doi.org/10.2174/1874450800802010038
Liu Z-W, Jarret RL, Kresovich S, Duncan RR (1995) Characterization and analysis of simple sequence repeat (SSR) loci in seashore paspalum (Paspalum vaginatum Swartz). Theor Appl Genet 91:47–52. https://doi.org/10.1007/BF00220857
Edwards A, Civitello A, Hammond HA, Caskey CT (1991) DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am J Hum Genet 49:746–756
Rosenberg NA, Burke T, Elo K et al (2001) Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics 159:699–713
Bérubé M, Danielsdottir AK, Aguilar A, et al (2006) High rates of gene flow among geographic locations in North Atlantic fin whales (Balaenoptera Physalus)
Rivera-León VE, Urbán J, Mizroch S et al (2019) Long-term isolation at a low effective population size greatly reduced genetic diversity in Gulf of California fin whales. Sci Rep 9:12391. https://doi.org/10.1038/s41598-019-48700-5
Bérubé M, Urbán J, Dizon AE et al (2002) Genetic identification of a small and highly isolated population of fin whales (Balaenoptera physalus) in the Sea of Cortez, Mexico. Conserv Genet 3:183–190
Bourret V, Macé M, Crouau-Roy B (2007) Genetic variation and population structure of western Mediterranean and northern Atlantic Stenella coeruleoalba populations inferred from microsatellite data. J Mar Biol Assoc UK 87:265–269. https://doi.org/10.1017/S0025315407054859
Engelhaupt D, Hoelzel AR, Nicholson C et al (2009) Female philopatry in coastal basins and male dispersion across the North Atlantic in a highly mobile marine species, the sperm whale (Physeter macrocephalus). Mol Ecol 18:4193–4205
Crouau-Roy B (1988) Genetic structure of cave-dwelling beetles populations: significant deficiencies of heterozygotes. Heredity 60:321–327. https://doi.org/10.1038/hdy.1988.49
Frasier TR, Rastogi T, Brown MW et al (2006) Characterization of tetranucleotide microsatellite loci and development and validation of multiplex reactions for the study of right whale species (genus Eubalaena). Mol Ecol Notes 6:1025–1029. https://doi.org/10.1111/j.1471-8286.2006.01417.x
Cunha HA, Watts PC (2007) Twelve microsatellite loci for marine and riverine tucuxi dolphins (Sotalia guianensis and Sotalia fluviatilis). Mol Ecol Notes 7:1229–1231. https://doi.org/10.1111/j.1471-8286.2007.01839.x
Perl RGB, Geffen E, Malka Y et al (2018) Population genetic analysis of the recently rediscovered Hula painted frog (Latonia nigriventer) reveals high genetic diversity and low inbreeding. Sci Rep 8:5588. https://doi.org/10.1038/s41598-018-23587-w
Panova M, Makinen T, Fokin M et al (2008) Microsatellite cross-species amplification in the genus Littorina and detection of null alleles in Littorina saxatilis. J Molluscan Stud 74:111–117. https://doi.org/10.1093/mollus/eym052
Primmer CR, Painter JN, Koskinen MT et al (2005) Factors affecting avian cross-species microsatellite amplification. J Avian Biol 36:348–360. https://doi.org/10.1111/j.0908-8857.2005.03465.x
Coughlan J, Mirimin L, Dillane E et al (2006) Isolation and characterization of novel microsatellite loci for the short-beaked common dolphin (Delphinus delphis) and cross-amplification in other cetacean species. Mol Ecol Notes 6:490–492. https://doi.org/10.1111/j.1471-8286.2006.01284.x
Chen L, Yang G (2009) A set of polymorphic dinucleotide and tetranucleotide microsatellite markers for the Indo-Pacific humpback dolphin (Sousa chinensis) and cross-amplification in other cetacean species. Conserv Genet 10:697–700. https://doi.org/10.1007/s10592-008-9618-x
Avise JC, Johns GC (1999) Proposal for a standardized temporal scheme of biological classification for extant species. Proc Natl Acad Sci 96:7358–7363. https://doi.org/10.1073/pnas.96.13.7358
Barbara T, Palma-Silva C, Paggi GM et al (2007) Cross-species transfer of nuclear microsatellite markers: potential and limitations. Mol Ecol 16:3759–3767. https://doi.org/10.1111/j.1365-294X.2007.03439.x
Kalinowski ST (2004) Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv Genet 5:539–543. https://doi.org/10.1023/B:COGE.0000041021.91777.1a
Slatkin M (1985) Rare alleles as indicators of gene flow. Evolution 39:53–65
Bérubé M, Palsbøll PJ (2018) Hybridism. In: Encyclopedia of marine mammals. Elsevier, Amsterdam, pp 496–501
Amaral AR, Lovewell G, Coelho MM et al (2014) Hybrid speciation in a marine mammal: the clymene dolphin (Stenella clymene). PLoS ONE 9:e83645. https://doi.org/10.1371/journal.pone.0083645
Fordyce RE (2018) Cetacean evolution. In: Encyclopedia of marine mammals. Elsevier, Amsterdam, pp 180–185
Sasaki T, Nikaido M, Hamilton H et al (2005) Mitochondrial phylogenetics and evolution of mysticete whales. Syst Biol 54:77–90. https://doi.org/10.1080/10635150590905939
Peyran C, Planes S, Tolou N et al (2020) Development of 26 highly polymorphic microsatellite markers for the highly endangered fan mussel Pinna nobilis and cross-species amplification. Mol Biol Rep. https://doi.org/10.1007/s11033-020-05338-1
Acknowledgements
We thank WWF-France for providing samples from Balaenoptera physalus, Physeter macrocephalus, Globicephala melas. We are also grateful to all the volunteers of WWF-France who participated in the campaigns at sea. We thank W. Mullié in Mauritania for the collection of Balaenoptera acutorostrata samples. Thank you to J. Almany for english corrections.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The sampling was non-lethal and a priori approved by the DREAL (Direction Régionale de l’Environnement, de l’Aménagement et du Logement) of Provence-Alpes Côte d’Azur.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tardy, C., Planes, S., Jung, JL. et al. Characterization of 25 new microsatellite markers for the fin whale (Balaenoptera physalus) and cross-species amplification in other cetaceans. Mol Biol Rep 47, 6983–6996 (2020). https://doi.org/10.1007/s11033-020-05757-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05757-0