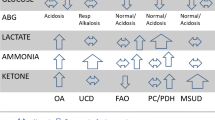
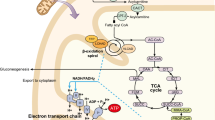
Abstract
Methylmalonic acidemia (MMA), an inherited metabolic disease, results from genetic defects in methylmalonyl-CoA mutase or any of the proteins involved in adenosylcobalamin synthesis. This enzyme is classified into several complementation groups and genotypic classes. In this work we explain the biochemical, structural and genetic analysis of 25 MMA patients, from Iran. The diagnosis was established by the measurement of propionylcarnitine in blood using tandem mass spectrometry and confirmed using a gas chromatography–flame ionization detector. Using clinical, biochemical, structural and molecular analyses we identified 15 mut MMA, three cblA, one cblB, and four cblC-deficient patients. Among mutations identified in the MUT gene (MUT) only one, the c.1874A>C (p.D625A) variant, is likely a mut− mutation. The remaining mutations are probably mut0. Here, we present the first molecular analysis of MMA in Iranian patients and have identified eight novel mutations. Four novel mutations (p.D625A, p.R326G, p.V157F, p.F379L) were seen exclusively in patients from northern Iran. One novel splice site mutation (c.2125-3C>G) in MUT and two novel mutation (p.N225M and p.A99P) in the MMAA gene were associated with patients from eastern Iran. The rs184829210 SNP was recognized only in patients with the novel c.958G>A (p.A320T) mutation. This study confirms pathogenesis of deficient enzyme activity in MUT, MMAA, MMAB, and MMACHC as previous observations. These results could act as a basis for the performance of pharmacological therapies for increasing the activity of proteins derived from these mutations.

Similar content being viewed by others
Abbreviations
- AdoCbl:
-
Adenosylcobalamin
- Cbl:
-
Cobalamin
- cDNA:
-
Complementary DNA
- GC–FID:
-
Gas chromatography–flame ionization detector
- HGMD:
-
Human gene mutation database
- MCM:
-
Methylmalonyl-CoA mutase
- MeCbl:
-
Methylcobalamin
- MMA:
-
Methylmalonic acidemia
- MSUD:
-
Maple syrup urine disease
- MUMS:
-
Mashhad University of Medical Sciences
- PA:
-
Propionic acidemia
- RT-PCR:
-
Reverse transcriptase-polymerase chain reaction
- SNP:
-
Single nucleotide polymorphism
References
Peters HL, Nefedov M, Lee LW et al (2002) Molecular studies in mutase-deficient (MUT) methylmalonic aciduria: identification of five novel mutations. Hum Mutat 20:406
Acquaviva C, Benoist JF, Pereira S et al (2005) Molecular basis of methylmalonyl-CoA mutase apoenzyme defect in 40 European patients affected by mut(o) and mut- forms of methylmalonic acidemia: identification of 29 novel mutations in the MUT gene. Hum Mutat 25:167–176
Martinez MA, Rincon A, Desviat LR, Merinero B, Ugarte M, Perez B (2005) Genetic analysis of three genes causing isolated methylmalonic acidemia: identification of 21 novel allelic variants. Mol Genet Metab 84:317–325
Rosenblatt DS, Fenton WA (2001) Inherited disorders of folate and cobalamin transport and metabolism. In: Scriver CR, Beaudet AL, Valle D, Sly WS (eds) The metabolic and molecular bases of inherited disease. McGraw-Hill, New York, pp 3897–3933
Fuchshuber A, Mucha B, Baumgartner ER, Vollmer M, Hildebrandt F (2000) mut0 methylmalonic acidemia: eleven novel mutations of the methylmalonyl CoA mutase including a deletion-insertion mutation. Hum Mutat 16:179
Jansen R, Ledley FD (1990) Heterozygous mutations at the mut locus in fibroblasts with mut0 methylmalonic acidemia identified by polymerase-chain-reaction cDNA cloning. Am J Hum Genet 47:808–814
Dobson CM, Wai T, Leclerc D et al (2002) Identification of the gene responsible for the cblA complementation group of vitamin B12-responsive methylmalonic acidemia based on analysis of prokaryotic gene arrangements. Proc Natl Acad Sci USA 99:15554–15559
Dobson CM, Wai T, Leclerc D et al (2002) Identification of the gene responsible for the cblB complementation group of vitamin B12-dependent methylmalonic aciduria. Hum Mol Genet 11:3361–3369
Lerner-Ellis JP, Anastasio N, Liu J et al (2009) Spectrum of mutations in MMACHC, allelic expression, and evidence for genotype-phenotype correlations. Hum Mutat 30:1072–1081
Richard E, Jorge-Finnigan A, Garcia-Villoria J et al (2009) Genetic and cellular studies of oxidative stress in methylmalonic aciduria (MMA) cobalamin deficiency type C (cblC) with homocystinuria (MMACHC). Hum Mutat 30:1558–1566
Han L, Gao X, Ye J, Qiu W, Gu X (2005) Application of tandem mass spectrometry in diagnosis of organic acidemias. Chin J Pediatr 43:325–330
Keyfi F, Varasteh A (2016) Development and validation of a GC-FID method for diagnosis of methylmalonic acidemia. Rep Biochem Mol Biol 4:104
Tanaka K, West-Dull A, Hine DG, Lynn TB, Lowe T (1980) Gas-chromatographic method of analysis for urinary organic acids. II. Description of the procedure, and its application to diagnosis of patients with organic acidurias. Clin Chem 26:1847–1853
Miller S, Dykes D, Polesky H (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215
Keyfi F, Sankian M, Moghaddassian M, Rolfs A, Varasteh A (2016) Molecular, biochemical, and structural analysis of a novel mutation in patients with methylmalonyl-CoA mutase deficiency. Gene 576:208–213
Froese DS, Kochan G, Muniz JR et al (2010) Structures of the human GTPase MMAA and vitamin B12-dependent methylmalonyl-CoA mutase and insight into their complex formation. J Biol Chem 285:38204–38213
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Weiner SJ, Kollman PA, Case DA et al (1984) A new force field for molecular mechanical simulation of nucleic acids and proteins. J Am Chem Soc 106:765–784
Li Z, Scheraga HA (1987) Monte Carlo-minimization approach to the multiple-minima problem in protein folding. Proc Natl Acad Sci USA 84:6611–6615
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Vriend G (1990) WHAT IF: a molecular modeling and drug design program. J Mol Graph 8:52–56
Keyfi F, Abbaszadegan MR, Rolfs A, Orolicki S, Moghaddassian M, Varasteh A (2016) Identification of a novel deletion in the MMAA gene in two Iranian siblings with vitamin B12-responsive methylmalonic acidemia. Cell Mol Biol Lett 21:4
Forny P, Froese DS, Suormala T, Yue WW, Baumgartner MR (2014) Functional characterization and categorization of missense mutations that cause methylmalonyl-CoA mutase (MUT) deficiency. Hum Mutat 35:1449–1458
Worgan LC, Niles K, Tirone JC et al (2006) Spectrum of mutations in mut methylmalonic acidemia and identification of a common Hispanic mutation and haplotype. Hum Mutat 27:31–43
Yang X, Sakamoto O, Matsubara Y et al (2004) Mutation analysis of the MMAA and MMAB genes in Japanese patients with vitamin B(12)-responsive methylmalonic acidemia: identification of a prevalent MMAA mutation. Mol Genet Metab 82:329–333
Lerner-Ellis JP, Tirone JC, Pawelek PD et al (2006) Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nat Genet 38:93–100
Ledley FD, Jansen R, Nham SU, Fenton WA, Rosenberg LE (1990) Mutation eliminating mitochondrial leader sequence of methylmalonyl-CoA mutase causes muto methylmalonic acidemia. Proc Natl Acad Sci USA 87:3147–3150
Raff ML, Crane AM, Jansen R, Ledley FD, Rosenblatt DS (1991) Genetic characterization of a MUT locus mutation discriminating heterogeneity in mut0 and mut- methylmalonic aciduria by interallelic complementation. J Clin Investig 87:203–207
Ogasawara M, Matsubara Y, Mikami H, Narisawa K (1994) Identification of two novel mutations in the methylmalonyl-CoA mutase gene with decreased levels of mutant mRNA in methylmalonic acidemia. Hum Mol Genet 3:867–872
Qureshi AA, Crane AM, Matiaszuk NV, Rezvani I, Ledley FD, Rosenblatt DS (1994) Cloning and expression of mutations demonstrating intragenic complementation in mut0 methylmalonic aciduria. J Clin Investig 93:1812–1819
Touraine RL, Rolland MO, Divry P, Mathieu M, Guibaud P, Bozon D (1995) A 13-bp deletion (1952 del 13) in the methylmalonyl CoA mutase gene of an affected patient. Hum Mutat 5:354–356
Adjalla CE, Hosack AR, Gilfix BM et al (1998) Seven novel mutations in mut methylmalonic aciduria. Hum Mutat 11:270–274
Champattanachai V, Ketudat Cairns JR, Shotelersuk V et al (2003) Novel mutations in a Thai patient with methylmalonic acidemia. Mol Genet Metab 79:300–302
Jung JW, Hwang IT, Park JE et al (2005) Mutation analysis of the MCM gene in Korean patients with MMA. Mol Genet Metab 84:367–370
Kobayashi A, Kakinuma H, Takahashi H (2006) Three novel and six common mutations in 11 patients with methylmalonic acidemia. Pediatr Int 48:1–4
Gradinger AB, Belair C, Worgan LC et al (2007) Atypical methylmalonic aciduria: frequency of mutations in the methylmalonyl CoA epimerase gene (MCEE). Hum Mutat 28:1045
Lempp TJ, Suormala T, Siegenthaler R et al (2007) Mutation and biochemical analysis of 19 probands with mut0 and 13 with mut- methylmalonic aciduria: identification of seven novel mutations. Mol Genet Metab 90:284–290
Filippi L, Gozzini E, Cavicchi C et al (2009) Insulin-resistant hyperglycaemia complicating neonatal onset of methylmalonic and propionic acidaemias. J Inherit Metab Dis 1:179–186
Perez B, Rincon A, Jorge-Finnigan A et al (2009) Pseudoexon exclusion by antisense therapy in methylmalonic aciduria (MMAuria). Hum Mutat 30:1676–1682
Yi Q, Lv J, Tian F, Wei H, Ning Q, Luo X (2011) Clinical characteristics and gene mutation analysis of methylmalonic aciduria. J Huazhong Univ Sci Technolog Med Sci 31:384–389
Dundar H, Ozgul RK, Guzel-Ozanturk A et al (2012) Microarray based mutational analysis of patients with methylmalonic acidemia: identification of 10 novel mutations. Mol Genet Metab 106:419–423
Liu MY, Liu TT, Yang YL et al (2012) Mutation profile of the MUT gene in Chinese methylmalonic aciduria patients. JIMD Rep 6:55–64
O’Shea CJ, Sloan JL, Wiggs EA et al (2012) Neurocognitive phenotype of isolated methylmalonic acidemia. Pediatrics 129:1541–1551
Vatanavicharn N, Champattanachai V, Liammongkolkul S et al (2012) Clinical and molecular findings in Thai patients with isolated methylmalonic acidemia. Mol Genet Metab 106:424–429
Crane AM, Jansen R, Andrews ER, Ledley FD (1992) Cloning and expression of a mutant methylmalonyl coenzyme A mutase with altered cobalamin affinity that causes mut- methylmalonic aciduria. J Clin Investig 89:385–391
Crane AM, Ledley FD (1994) Clustering of mutations in methylmalonyl CoA mutase associated with mut- methylmalonic acidemia. Am J Hum Genet 55:42–50
Janata J, Kogekar N, Fenton WA (1997) Expression and kinetic characterization of methylmalonyl-CoA mutase from patients with the mut- phenotype: evidence for naturally occurring interallelic complementation. Hum Mol Genet 6:1457–1464
Ledley FD, Rosenblatt DS (1997) Mutations in mut methylmalonic acidemia: clinical and enzymatic correlations. Hum Mutat 9:1–6
Benoist JF, Acquaviva C, Callebaut I et al (2001) Molecular and structural analysis of two novel mutations in a patient with mut(-) methylmalonyl-CoA deficiency. Mol Genet Metab 72:181–184
Ostergaard E, Wibrand F, Orngreen MC, Vissing J, Horn N (2005) Impaired energy metabolism and abnormal muscle histology in mut- methylmalonic aciduria. Neurology 65:931–933
Lerner-Ellis JP, Dobson CM, Wai T et al (2004) Mutations in the MMAA gene in patients with the cblA disorder of vitamin B12 metabolism. Hum Mutat 24:509–516
Merinero B, Perez B, Perez-Cerda C et al (2008) Methylmalonic acidaemia: examination of genotype and biochemical data in 32 patients belonging to mut, cblA or cblB complementation group. J Inherit Metab Dis 31:55–66
Guven A, Cebeci N, Dursun A, Aktekin E, Baumgartner M, Fowler B (2012) Methylmalonic acidemia mimicking diabetic ketoacidosis in an infant. Pediatr Diabetes 13:22–25
Dempsey-Nunez L, Illson ML, Kent J et al (2012) High resolution melting analysis of the MMAA gene in patients with cblA and in those with undiagnosed methylmalonic aciduria. Mol Genet Metab 107:363–367
Lerner-Ellis JP, Gradinger AB, Watkins D et al (2006) Mutation and biochemical analysis of patients belonging to the cblB complementation class of vitamin B12-dependent methylmalonic aciduria. Mol Genet Metab 87:219–225
Keeratichamroen S, Cairns JR, Sawangareetrakul P et al (2007) Novel mutations found in two genes of thai patients with isolated methylmalonic acidemia. Biochem Genet 45:421–430
Jorge-Finnigan A, Aguado C, Sanchez-Alcudia R et al (2010) Functional and structural analysis of five mutations identified in methylmalonic aciduria cblB type. Hum Mutat 31:1033–1042
Heil SG, Hogeveen M, Kluijtmans LA et al (2007) Marfanoid features in a child with combined methylmalonic aciduria and homocystinuria (CblC type). J Inherit Metab Dis 30:811
Yuen YP, Lai CK, Chan YW, Lam CW, Tong SF, Chan KY (2007) DNA-based diagnosis of methylmalonic aciduria and homocystinuria, cblC type in a Chinese patient presenting with mild developmental delay. Clin Chim Acta 375:171–172
Nogueira C, Aiello C, Cerone R et al (2008) Spectrum of MMACHC mutations in Italian and Portuguese patients with combined methylmalonic aciduria and homocystinuria, cblC type. Mol Genet Metab 93:475–480
Bouts AH, Roofthooft MT, Salomons GS, Davin JC (2010) CD46-associated atypical hemolytic uremic syndrome with uncommon course caused by cblC deficiency. Pediatr Nephrol 25:2547–2548
Liu MY, Yang YL, Chang YC et al (2010) Mutation spectrum of MMACHC in Chinese patients with combined methylmalonic aciduria and homocystinuria. J Hum Genet 55:621–626
Wang F, Han L, Yang Y et al (2010) Clinical, biochemical, and molecular analysis of combined methylmalonic acidemia and hyperhomocysteinemia (cblC type) in China. J Inherit Metab Dis 3:435–442
Sakamoto O, Ohura T, Matsubara Y, Takayanagi M, Tsuchiya S (2007) Mutation and haplotype analyses of the MUT gene in Japanese patients with methylmalonic acidemia. J Hum Genet 52:48–55
Berger I, Shaag A, Anikster Y et al (2001) Mutation analysis of the MCM gene in Israeli patients with mut(0) disease. Mol Genet Metab 73:107–110
Tollinger M, Konrat R, Hilbert BH, Marsh ENG, Kräutler B (1998) How a protein prepares for B 12 binding: structure and dynamics of the B 12-binding subunit of glutamate mutase from Clostridium tetanomorphum. Structure 6:1021–1033
Cervantes M, Murillo FJ (2002) Role for vitamin B12 in light induction of gene expression in the bacterium Myxococcus xanthus. J Bacteriol 184:2215–2224
Acknowledgements
This research was supported and funded by the Immunology Research Center, School of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare they have no conflicts of interest.
Additional information
Accession Numbers: The accession number for the c.1874A>C, p.D625A reported in this paper is [NCBI]: [KP289323]. The accession number for the c.976A>G/N, p.R326G reported in this paper is [NCBI]: [KP289324]. The accession number for the c.469G>T, p. V157F reported in this paper is [NCBI]: [KR026956]. The accession number for the c.2125-3C>G reported in this paper is [NCBI]: [KF030882]. The accession number for the c.1137del T, p.F379L reported in this paper is [NCBI]: [KR026957]. The accession number for the c.674del A, p.N225M reported in this paper is [NCBI]: [KR026958].
Electronic supplementary material
Below is the link to the electronic supplementary material.

11033_2018_4469_MOESM1_ESM.jpg
Supplementary Figure 1 Three-dimensional structures of normal and mutant MUT proteins with novel mutation (D625A). Whole view of the mutant protein (Left Above), close-up view of the mutant protein (Left Below), whole view of the normal protein (Right Above), and close-up view of the normal protein (Right Below). (JPG 254 KB)

11033_2018_4469_MOESM2_ESM.jpg
Supplementary Figure 2 Three-dimensional structures of normal and mutant MUT proteins with novel mutation (R326G). Whole view of the mutant protein (Left Above), close-up view of the mutant protein (Left Below), whole view of the normal protein (Right Above), and close-up view of the normal protein (Right Below). (JPG 250 KB)

11033_2018_4469_MOESM3_ESM.jpg
Supplementary Figure 3 Three-dimensional structures of normal and mutant MUT proteins with novel mutation (V157F). Whole view of the mutant protein (Left Above), close-up view of the mutant protein (Left Below), whole view of the normal protein (Right Above), and close-up view of the normal protein (Right Below). (JPG 230 KB)

11033_2018_4469_MOESM4_ESM.jpg
Supplementary Figure 4 Three-dimensional structures of normal and mutant MUT proteins representing novel mutation (A320T), whole view of the mutant protein (Left Above), close-up view of the mutant protein (Left Below), whole view of the normal protein (Right Above), and close-up view of the normal protein (Right Below). (JPG 252 KB)
Rights and permissions
About this article
Cite this article
Keyfi, F., Abbaszadegan, M.R., Sankian, M. et al. Mutation analysis of genes related to methylmalonic acidemia: identification of eight novel mutations. Mol Biol Rep 46, 271–285 (2019). https://doi.org/10.1007/s11033-018-4469-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4469-0