Abstract
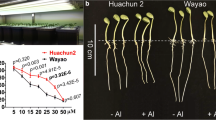
Development of aluminium (Al) resistant genotypes through molecular breeding is a major approach for increasing seed yield under acidic conditions. There are no available reports on mapping of Al resistance loci and molecular breeding for Al resistant varieties in lentil. The present study reports a major quantitative trait loci (QTL) for Al resistance using simple sequence repeat (SSR) markers in F2 and F3 mapping populations derived from contrasting parents. Phenotypic response to Al was measured on the bases of root re-growth (RRG), fluorescent signals (callose accumulation) and Al contents in hydroponic assay. After screening 495 SSR markers to search polymorphism between two contrasting parents, 73 polymorphic markers were used for bulk segregation analysis. Two major QTLs were identified using seven trait linked markers, one each for fluorescent signals and RRG mapped on linkage group (LG) 1 under Al stress conditions in F2 mapping population of cross BM-4 × L-4602. One major QTL (qAlt_fs) was localised between PLC_88 and PBA_LC_373, covering 25.9 cM with adjacent marker PLC_88 at a distance of 0.4 cM. Another major QTL (qAlt_rrg) for RRG was in the marker interval of PBA_LC_1247 and PLC_51, covering a distance of 45.7 cM with nearest marker PBA_LC_1247 at a distance of 21.2 cM. Similarly, in F3 families of BM-4 × L-4602 and BM-4 × L-7903, LG-1 was extended to 285.9 and 216.4 cM respectively, having four newly developed genic-SSR markers. These QTLs had a logarithm of odd (LOD) value of 140.5 and 28.8 along with phenotypic variation of 52% and 11% for fluorescent signals and RRG respectively, whereas, qAlt_rrg had LOD of 36 and phenotypic variance of 25% in F3 population of BM-4 × L-4602. Two major QTLs identified in the present study can be further dissected for candidate gene discovery and development of molecular markers for breeding improved varieties with high Al resistance.




Similar content being viewed by others

References
Von Uexküll HR, Mutert E (1995) Global extent, development and economic impact of acid soils. Plant soil 171(1):1–15
Kochian LV (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Biol 46:237–260
Wissemeier AH, Klotz F, Horst WJ (1987) Aluminium induced callose synthesis in roots of soybean (Glycine max L). J Plant Physiol 129:487–492
Zhang G, Hoddinott J, Taylor GJ (1994) Characterization of 1,3-β-D-glucan (callose) synthesis in roots of Triticum aestivum in response to aluminium toxicity. J Plant Physiol 144:229–234
Singh D, Dikshit HK, Kumar A (2015) Aluminium tolerance in lentil with monogenic inheritance pattern. Plant Breed 134:105–110
Singh D, Pal M, Singh CK, Jyoti T, Jain P, Chaturvedi AK, Sadhana M, Karwa S, Singh R, Tomar RSS, Rita N, Nandini C, Singh MP (2016) Molecular scanning and morpho-physiological dissection of component mechanism in Lens species in response to aluminium stress. PLoS ONE. https://doi.org/10.1371/journal.pone.0160073.
Horst WJ, Puschel AK, Schmohl N (1997) Induction of callose formation is a sensitive marker for genotypic aluminium sensitivity in maize. Plant Soil 192:23–30
Tesfaye M, Temple SJ, Allan DL, Vance CP, Samac DA (2001) Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol 127(4):1836–1844
Singh D, Chauhan SK (2011) Organic acids of crop plants in aluminium detoxification. Curr Sci 100:1509–1515
Marschner H (1986) Mineral nutrition of higher plants. Academic Press, London
Miyasaka SC, Kochian LV, Shaff J, Foy CD (1989) Mechanisms of aluminum tolerance in wheat: an investigation of genotypic differences in rhizosphere pH, K+, and H+ transport, and root-cell membrane potentials. Plant Physiol 91(3):1188–1196
Taylor GJ (1995) Overcoming barriers to understanding the cellular basis of aluminium resistance. Plant Soil 171(1):89–103
Degenhardt J, Larsen PB, Howell SH, Kochian LV (1998) Aluminum resistance in the Arabidopsis mutantalr-104 is caused by an aluminum-induced increase in rhizosphere pH. Plant Physiol 117(1):19–27
Kinraide TB, Parker DR, Zobel RW (2005) Organic acid secretion as a mechanism of aluminium resistance: a model incorporating the root cortex, epidermis, and the external unstirred layer. J Exp Bot 56(417):1853–1865
Basu A, Basu U, Taylor GJ (1994) Induction of microsomal membrane proteins in roots of an aluminium resistant cultivar of Triticum aestivum L. under conditions of aluminium stress. Plant Physiol 104:1007–1013
Jones DL, Brassington DS (1998) Sorption of organic acids in acid soils and its implications in the rhizosphere. Eur J Soil Sci 49:447–455
Singh D, Dikshit HK, Singh R (2012) Variation of aluminium tolerance in lentil. Plant Breed 131:751–761
Singh D, Raje RS (2011) Genetics of aluminium tolerance in chickpea. Plant Breed 130(5):563–568
Singh D, Raje RS, Choudhary AK (2011) Genetic control of aluminium tolerance in pigeonpea. Crop Pasture Sci 62:761–764
Singh D, Choudhary AK (2010) Inheritance of aluminium tolerance in pea. Plant Breed 129:688–692
Horst WJ (1987) Aluminium tolerance and calcium efficiency of cowpea genotypes. J Plant Nutr 10:9–16
Raman H, Moroni JS, Sato K, Read BJ, Scott BJ (2002) Identification of AFLP and microsatellite markers linked with an aluminium tolerance gene in barley (Hordeum vulgare L.). Theor Appl Genet 105:458–464
Raman H, Zhang K, Cakir M, Appels R, Garvin DF, Maron LG, Kochian LV, Moroni JS, Raman R, Imtiaz M, Drake-Brockman F (2005) Molecular characterization and mapping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48(5):781–791
Ma JF, Shen R, Zhao Z, Wissuwa M, Takeuchi Y et al (2002) Response of rice to Al stress and identification of quantitative trait loci for Al tolerance. Plant Cell Physiol 43:652–659
Zhou LL, Bai GH, Ma HX, Carver BF (2007) Quantitative trait loci for aluminium resistance in wheat. Mol Breed 19:153–161
Guimaraes CT, Simoes CC, Pastina MM, Maron LG, Magalhaes JV et al (2014) Genetic dissection of Al tolerance QTls in the maize genome by high density SNP scan. BMC Genomics 15:153–167
Navabi A, Mather DE, Bernier J, Spaner DM, Atlin GN (2009) QTL detection with bidirectional and unidirectional selective genotyping: marker-based and trait-based analyses. Theor Appl Genet 118:347–358
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Subudhi PK, Borkakati RP, Virmani SS, Huang N (1997) Molecular mapping of a thermosensitive genetic male sterility gene in rice using bulked segregant analysis. Genome 40(2):188–194. https://doi.org/10.1139/g97-027
Xue Y, Jiang L, Su N, Wang JK, Deng P, Ma JF, Zhai HQ, Wan JM (2007) The genetic basic and Wne-mapping of a stable quantitative-trait loci for aluminium tolerance in rice. Planta 227:255–262
Ma Y, Li C, Ryan PR, Shabala S, You J, Liu J, Liu C, Zhou M (2016) A new allele for aluminium tolerance gene in barley (Hordeum vulgare L.). BMC Genomics 17(1):186
Ninamango-Cárdenas FE, Guimarães CT, Martins PR, Parentoni SN, Carneiro NP, Lopes MA, Moro JR, Paiva E (2003) Mapping QTLs for aluminum tolerance in maize. Euphytica 130:223–232
Polle E, Konzak CF, Kittrick JA (1978) Visual detection of aluminium tolerance levels in wheat by hematoxylin staining of seedlings roots. Crop Sci 18:823–827
Kauss H (1992) Callose and callose synthase. In: Gurr SJ, McPherson MJ, Bowles DJ (eds) Molecular plant pathology: a practical approach, vol 2. Oxford University Press, New York, pp 1–8
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Hamwieh A, Udupa SM, Choumane W, Sarker A, Dreyer F, Jung C et al (2005) A genetic linkage map of Lens sp. based on microsatellite and AFLP markers and the localization of Fusarium vascular wilt resistance. Theor Appl Genet 110:669–677
Kaur S, Cogan N, Amber S, Noy D, Butsch M, Froster JW et al (2014) EST-SNP discovery and dense genetic mapping in lentil (Lens culinaris Medik.) enable candidate gene selection for boron tolerance. Theor Appl Genet 127(3):703–713
Jain N, Dikshit HK, Singh D, Singh A, Kumar H (2013) Discovery of EST-derived microsatellite primers in the legume Lens culinaris (Fabaceae). Appl Plant Sci. https://doi.org/10.3732/apps.1200539
Singh D, Singh CK, Taunk J, Chaturvedi SK, Kishor G, Pal M (2017) Transcriptome analysis of lentil (Lens culinaris Medikus) in response to seedling drought stress. BMC Genomics 18:206. https://doi.org/10.1186/s12864-017-3596-7
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1(2):174–181
Kosambi DD (1994) The estimation of a map distance from recombination values. Ann Eugen 12(3):172–175
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
Wang B, Porter AH (2004) An AFLP-based interspecific linkage map of sympatric, hybrid Colias butterflies. Genetics 168(1):215–225
Rao IM, Miles JW, Beebe SE, Horst WJ (2016) Root adaptations to soils with low fertility and aluminium toxicity. Ann Bot 118(4):593–605. https://doi.org/10.1093/aob/mcw073
Baier AC, Somers DJ, Gusiafson JP (1995) Aluminium tolerance in wheat: correlating hydroponic evaluations with field and soil performances. Plant Breed 114:291–296. https://doi.org/10.1111/j.1439-0523.1995.tb01236.x
Singh D, Pal M, Singh R, Singh CK, Chaturvedi AK (2015) Physiological and biochemical characteristics of Vigna species for Al stress tolerance. Acta Physiol Plant 37(4):87
Wang J, Raman H, Zhou M, Ryan PR, Delhaize E, Hebb DM, Coombes N, Mendham N (2007) High-resolution mapping of the Alp locus and identification of a candidate gene HvMATE controlling aluminium tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 115:265–276
Navakode S, Weidner A, Lohwasser U, Roder MS, Borner A (2009) Molecular mapping of quantitative trait loci (QTLs) controlling aluminium tolerance in bread wheat. Euphytica 166(2):283–290
Pirselova B, Matusikova I (2013) Callose: the plant cell wall polysaccharide with multiple biological functions. Acta Physiol Plant 35(3):635–644
Zhang H, Shi WL, You JF, Bian MD, Qin XM. Yu H, Liu Q, Peter R, Yang ZM (2015) Transgenic Arabidopsis thaliana plants expressing a β-1,3-glucanase from sweet sorghum (Sorghum bicolor L.) show reduced callose deposition and increased tolerance to aluminium toxicity. Plant Cell Environ 38:1178–1188
Too EJ, Carlsson AS, Onkware AO et al (2014) Cell membrane integrity, callose accumulation, and root growth in aluminum-stressed sorghum seedlings. Biol Plant 58:768. https://doi.org/10.1007/s10535-014-0455-0
Alvim MN, Ramos FT, Oliveira DC, Isaias RMS, França MGC (2012) Aluminium localization and toxicity symptoms related to root growth inhibition in rice (Oryza sativa L.) seedlings. J Biosci 37(6):1079–1088
Stass A, Wang Y, Eticha D, Horst WJ (2006) Aluminium rhizotoxicity in maize grown in solutions with Al3+ or Al(OH)4 − as predominant solution Al species. J Exp Bot 57(15):4033–4042
Soto-Cerda BJ, Inostroza-Blancheteau C, Mathias M, Penaloza E, Zuniga J, Munoz G et al (2015) Marker-assisted breeding for TaALMT1, a major gene conferring aluminium tolerance to wheat. Biol Plant 59:83–91
Camargo CE, Ferreira Filho AW (2005) Genetic control of wheat seedling root growth. Sci Agric 62(4):325–330
Nguyen VT, Burow MD, Nguyen HT, Le BT, Le TD, Paterson AH (2001) Molecular mapping of genes conferring aluminum tolerance in rice (Oryza sativa L.). Theor Appl Genet 102(6–7):1002–1010
Magalhaes JV, Garvin DF, Wang Y, Sorrells ME, Klein PE, Schaffert RE, Li L, Kochian LV (2004) Comparative mapping of a major aluminum tolerance gene in sorghum and other species in the Poaceae. Genetics 167(4):1905–1914
Ma HX, Bai GH, Carver BF, Zhou LL (2005) Molecular mapping of a quantitative trait locus for aluminum tolerance in wheat cultivar Atlas 66. Theor Appl Genet 112(1):51
Riede CR, Anderson JA (1995) Linkage of RFLP markers to an aluminum tolerance gene in wheat. Crop Sci 36(4):905–909
Acknowledgements
Authors thank to Director, Joint Director (Res), ICAR-Indian Agricultural Research Institute (IARI), New Delhi, Head, Division of Genetics and Incharge, National Phytotron Facility, IARI, New Delhi, for given support to accomplish the research activities. The financial support from ICAR-IARI (Project no-JAN 09/16), New Delhi is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: DS, MP, KCU; Performed the experiments: CKS, SK; Analyzed the data: CKS, RSST; Drafted the manuscript: DS, CKS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no competing interests.
Research involving human and animal rights
This article does not contain any studies with human participants and/or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Determination of Al contents in roots of tolerant parent and tolerant F2 plants as well as sensitive parent and sensitive F2 plants. (TIF 10212 KB)

Fig. S2
Electrophoretic profile of PLC_88 in parents, bulks and selected F2 individuals. TP- Tolerant parent (L-4602), SP- sensitive parent (BM-4), TB- tolerant bulk, SB- Sensitive bulk, L- 100bp Ladder. (JPG 111 KB)

Fig. S3
Validation of Al linked marker, PLC_88 in Al resistant and sensitive genotypes. T- Tolerant parent, S- Sensitive parent of F2 population. (JPG 220 KB)
Rights and permissions
About this article
Cite this article
Singh, C.K., Singh, D., Tomar, R.S.S. et al. Molecular mapping of aluminium resistance loci based on root re-growth and Al-induced fluorescent signals (callose accumulation) in lentil (Lens culinaris Medikus). Mol Biol Rep 45, 2103–2113 (2018). https://doi.org/10.1007/s11033-018-4368-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4368-4