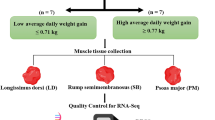
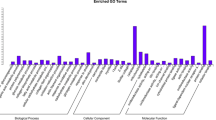
Abstract
Chianina and Maremmana breeds play an important role in the Italian cattle meat market. The Chianina breed is an ancient breed principally raised for draught. Now this breed is the worldwide recognized producer of top quality beef, tasteful and tender, specifically the famous “Florentine steak”. The Maremmana characterized by a massive skeletal structure, is a rustic cattle breed selected for adaptability to the marshy land of the Maremma region. We used a high throughput mRNA sequencing to analyze gene expression in muscle tissues of two Italian cattle breeds, Maremmana (MM) and Chianina (CN) with different selection history. We aim to examine the specific genetic contribution of each breed to meat production and quality, comparing the skeletal muscle tissue from Maremmana and Chianina. Most of the differentially expressed genes were grouped in the Glycolysis/Gluconeogenesis pathways. The rate and the extent of post-mortem energy metabolism have a critical effect on the conversion of muscle to meat. Furthermore, we aim at discovering the differences in nucleotide variation between the two breeds which might be attributable to the different history of selection/divergence. In this work we could emphasize the involvement of pathways of post-mortem energy metabolism. Moreover, we detected a collection of coding SNPs which could offer new genomic resources to improve phenotypic selection in livestock breeding program.





Similar content being viewed by others
Abbreviations
- MM:
-
Maremmana
- CN:
-
Chianina
- SNP:
-
Single nucleotide polymorphism
- FPKM:
-
Fragments per kilobase of exon per million fragments mapped
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- DEG:
-
Differentially expressed gene
- ES:
-
Exon skipping
- A5SS:
-
Alternative 5′ splice site
- A3SS:
-
Alternative 3′ splice site
- RI:
-
Retained intron
- OT:
-
Others
- TSS:
-
Single transcription start site
- UTR:
-
Untraslated region
- SIFT:
-
Sorting intolerant from tolerant
References
Cozzi G, Ragno E (2003) Meat production and market in Italy. Agric Conspec Sci 68:7
Pellecchia M, Negrini R, Colli L, Patrini M, Milanesi E, Achilli A, Bertorelle G, Cavalli-Sforza LL, Piazza A, Torroni A et al (2007) The mystery of Etruscan origins: novel clues from Bos taurus mitochondrial DNA. Proc Biol Sci 274:1175–1179
Negrini R, Milanesi E, Bozzi R, Pellecchia M, Ajmone-Marsan P (2006) Tuscany autochthonous cattle breeds: an original genetic resource investigated by AFLP markers. J Anim Breed Genet 123:10–16
Felius M (1995) Cattle breeds—an encyclopedia, 1st edn. Misset, Doetinchem
Bongiorni S, Gruber CEM, Bueno S, Chillemi G, Ferrè F, Failla S, Moioli B, Valentini A (2016) Transcriptomic investigation of meat tenderness in two Italian cattle breeds. Anim Genet. doi:10.1111/age.12418
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Roberts A, Pimentel H, Trapnell C, Pachter L (2011) Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27:2325–2329
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:11
Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576
Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:P3
Maere S, Heymans K, Kuiper M (2005) BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Rosado M, Barber CF, Berciu C, Feldman S, Birren SJ, Nicastro D, Goode BL (2014) Critical roles for multiple formins during cardiac myofibril development and repair. Mol Biol Cell 25:811–827
Wei Z, Sun M, Liu X, Zhang J, Jin Y (2014) Rufy3, a protein specifically expressed in neurons, interacts with actin-bundling protein Fascin to control the growth of axons. J Neurochem 130:678–692
Li X, Baumgart E, Dong GX, Morrell JC, Jimenez-Sanchez G, Valle D, Smith KD, Gould SJ (2002) PEX11alpha is required for peroxisome proliferation in response to 4-phenylbutyrate but is dispensable for peroxisome proliferator-activated receptor alpha-mediated peroxisome proliferation. Mol Cell Biol 22:8226–8240
Wheeler TL, Cundiff LV, Shackelford SD, Koohmaraie M (2004) Characterization of biological types of cattle (Cycle VI): carcass, yield, and longissimus palatability traits. J Anim Sci 82:1177–1189
Dunner S, Sevane N, Garcia D, Levéziel H, Williams JL, Mangin B, Valentini A, GeMQual Consortium (2013) Genes involved in muscle lipid composition in 15 European Bos taurus breeds. Anim Genet 44:493–501
Pariset L, Mariotti M, Nardone A, Soysal MI, Ozkan E, Williams JL, Dunner S, Leveziel H, Maróti-Agóts A, Bodò I et al (2010) Relationships between Podolic cattle breeds assessed by single nucleotide polymorphisms (SNPs) genotyping. J Anim Breed Genet 127:481–488
Blott SC, Williams JL, Haley CS (1999) Discriminating among cattle breeds using genetic markers. Heredity (Edinb) 82(Pt 6):613–619
Hwang IH, Park BY, Kim JH, Cho SH, Lee JM (2005) Assessment of postmortem proteolysis by gel-based proteome analysis and its relationship to meat quality traits in pig longissimus. Meat Sci 69:79–91
Gutiérrez-Gil B, Wiener P, Nute GR, Burton D, Gill JL, Wood JD, Williams JL (2008) Detection of quantitative trait loci for meat quality traits in cattle. Anim Genet 39:51–61
Timperio AM, D’Alessandro A, Pariset L, D’Amici GM, Valentini A, Zolla L (2009) Comparative proteomics and transcriptomics analyses of livers from two different Bos taurus breeds: “Chianina and Holstein Friesian”. J Proteomics 73:309–322
Koohmaraie M, Geesink GH (2006) Contribution of postmortem muscle biochemistry to the delivery of consistent meat quality with particular focus on the calpain system. Meat Sci 74:34–43
Ferguson DM, Daly BL, Gardner GE, Tume RK (2008) Effect of glycogen concentration and form on the response to electrical stimulation and rate of post-mortem glycolysis in ovine muscle. Meat Sci 78:202–210
Du M, Shen QW, Zhu MJ (2005) Role of beta-adrenoceptor signaling and AMP-activated protein kinase in glycolysis of postmortem skeletal muscle. J Agric Food Chem 53:3235–3239
Cheung PC, Salt IP, Davies SP, Hardie DG, Carling D (2000) Characterization of AMP-activated protein kinase gamma-subunit isoforms and their role in AMP binding. Biochem J 346(Pt 3):659–669
Costford SR, Kavaslar N, Ahituv N, Chaudhry SN, Schackwitz WS, Dent R, Pennacchio LA, McPherson R, Harper ME (2007) Gain-of-function R225 W mutation in human AMPKgamma(3) causing increased glycogen and decreased triglyceride in skeletal muscle. PLoS One 2:e903
Milan D, Jeon JT, Looft C, Amarger V, Robic A, Thelander M, Rogel-Gaillard C, Paul S, Iannuccelli N, Rask L et al (2000) A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science 288:1248–1251
Berg M, Tymoczko JL, Stryer L (2002) Glycolysis and Gluconeogenesis (Ch. 16). In Biochemistry, 5th edn. W.H. Freeman, New York
Acknowledgments
This manuscript is dedicated to the memory of our friend and colleague Lorraine Pariset. Her work and passion on livestock genetics and her inquiring mind will be always an inspiration to us. This work is part of the GENZOOT research program, funded by Italian Ministry of Agricultural, Forestry and Food Policies (MIPAAF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics statement
This study was carried out in strict accordance with the European Union recommendation directive 2010/63/EU and the Italian low 116/92 about animal care.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bongiorni, S., Gruber, C.E.M., Chillemi, G. et al. Skeletal muscle transcriptional profiles in two Italian beef breeds, Chianina and Maremmana, reveal breed specific variation. Mol Biol Rep 43, 253–268 (2016). https://doi.org/10.1007/s11033-016-3957-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-3957-3