Abstract
Currently, the molecular regulation mechanisms of disease-resistant involved in maize leaf sheaths infected by banded leaf and sheath blight (BLSB) are poorly known. To gain insight into the transcriptome dynamics that are associated with their disease-resistant, genome-wide gene expression profiling was conducted by Solexa sequencing. More than four million tags were generated from sheath tissues without any leaf or development leaf, including 193,222 and 204,824 clean tags in the two libraries, respectively. Of these, 82,864 (55.4 %) and 91,678 (51.5 %) tags were matched to the reference genes. The most differentially expressed tags with log2 ratio >2 or <−2 (P < 0.001) were further analyzed, representing 1,476 up-regulated and 1,754 down-regulated genes, except for unknown transcripts, which were classified into 11 functional categories. The most enriched categories were those of metabolism, signal transduction and cellular transport. Next, the expression patterns of 12 genes were assessed by quantitative real-time PCR, and it is showed the results were general agreement with the Solexa analysis, although the degree of change was lower in amplitude. In conclusion, we first reveal the complex changes in the transcriptome during the early development of maize sheath infected by BLSB and provide a comprehensive set of data that are essential for understanding its molecular regulation mechanism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rhizoctonia solani Kühn is a plant pathogenic fungus with a wide host range and worldwide distribution and has a significant economic impact in the development and production of a wide variety of crops. To date, 13 Anastomosis Groups (AGs) have been recognized according to hyphal anastomosis behavior, cultural morphology, host range and pathogenicity [54]. Furthermore, the variability in the degree of hyphal fusion, morphology, pathogenicity and host range observed in AGs (AG-1, AG-13) and AG-1A has been widely endanger to maize as a dominant fungus in southwest of China and identified as anastomosis group [60].
Banded leaf and sheath blight (BLSB), caused by R. solani Kühn, is one of the most important diseases of maize (Zea mays.) worldwide. Severe yield losses can result as a consequence of the rapid development and large-scale spread of the disease epidemic under optimal environmental conditions. Futhermore, R. solani Kühn is a common soil borne pathogen with a great diversity of host plants that can attack previously resistant cultivars, along with the capacity of fungal spores to travel long distances, making control of BLSB difficult [61]. And this disease initially infects maize at the first and second leaf sheath above the ground and then spreads upward to infect the ear, leading to severe yield losses [61]. Recently, yield losses approached 100 % in southern of China when the ear rot phase predominated [27].
Over the last few years, genetic and molecular studied on the disease and pathogen have been reported in maize [36, 39, 62]. These studies have revealed that resistance to BLSB is a typical quantitative trait controlled by polygenes and three significant QTL located on chromosomes two, six, and ten to be responsible for resistance to BLSB respectively [8, 13, 61]. In addition, many catalytic enzymes involved in response to BLSB infection were analyzed, including chitinase, glucanase and phenylanine ammonia lyase [32, 35] and few pathogenesis-associated genes and some potential defense pathways were involved in response to BLSB infection [2, 64]. However, the use of most genetic and molecular techniques in studying genes involved in the maize-BLSB interactions has been largely limited due to the BLSB that is hard to produce any asexual (conidia) spores although it is considered to have an asexual life cycle.
To understand the mechanism(s) of the host resistance at the molecular level, a critical first step is to identify the transcripts that accumulate in response to the pathogen attack. In this study, “R15”, a identified inbred lines with good general combining ability, good agronomic characteristics and high level of resistance to BLSB [59], was employed to identify a set of candidate genes associated with BLSB resistance using Solexa’s Digital Gene Expression (DGE) technology. Which is upgraded from the previous massively parallel signature sequencing (MPSS) technology, [7, 29, 42] can produce the specific 3′signature for each mRNA, thereby reducing library saturation from abundant transcripts and enhancing the capacity for rare transcript detection [4, 43]. This study will help to elucidate the molecular mechanism of resistance to BLSB and provide important evidence for breeding excellent maize lines.
Materials and methods
Plants material and pathogen infection
High-resistance maize inbred line seedlings of “R15” were treated with 7 % hypochlorite solution for 30 min respectively, followed by three washes with sterilized water before being sowed in pots with autoclaved soil. Control plants were maintained under the same conditions. R. solani AG1-IA was cultured on potato dextrose agar and incubated at 28 °C for three days before use (Support information Figure S1). Infections were conducted by inoculating with R. solani (kindly provided by the Rice Institution of Sichuan Agricultural University, Sichuan, China) at 28 °C after growing for 35 days. The leave sheaths were covered with plastic bags to ensure high humidity. The leaf sheaths without any leaves were harvested from each of three maize plants, and the three leaf sheaths were combined to represent one replicate. Three independent replicates were collected for each sample. Infected leave sheaths were collected every 12 h for 2 days. Control samples were harvested from water-treated leaf sheaths incubated under the same conditions. (Support information Figure S2).
Preparation of digital expression libraries
Samples from infected leaf sheaths from 36 to 48 h were pooled for RNA isolation and library construction. Comparable control leaf sheaths were treated identically and in parallel. Sequence tag preparation was done with the Digital Gene Expression Tag Profiling Kit according to the manufacturer’s protocol. 20 μg of total RNA and 6 μg of mRNA were obtained and purified by adsorption of biotin Oligo(dT) magnetic beads. After mRNA’s binding, cDNA synthesis was performed. Double strand cDNA was introduced into cDNA fragment digested by NlaIII endonuclease and these binging fragment containing sequences of CATG site and adjacent polyA tail in 3′ end. These 3′ cDNA fragments were purified using magnetic bead precipitation and the Illumina adapter 1 (GEX adapter 1) was added to new 5′ end. The junction of Illumina adapter 1 and CATG site was recognized by MmeI, which cut at downstream CATG site and produce fragment of 17 bp tags with adaptor 1. After removing 3′ fragments with magnetic bead precipitation, the Illumina adapter 2 (GEX adapter 2) was ligated to 3′ end of the cDNA tag. These cDNA fragments represented the tag library. After denaturation, the single chain molecules are fixed onto the Illumina sequencing chip (flow cell). Then these sequences were prepared for Solexa sequencing.
Solexa sequencing
Sequencing was performed by Beijing Genomic institution. PCR phusion amplification with 15 cycles was performed with primers complementary to the adapter sequences to enrich the samples for the desired fragments. The 85 base strips were selected and purified by 6 % TBE PAGE Gel electrophoresis, and then digested and the single-chain molecules were fixed onto the Solexa Sequencing Chip (flow cell). Four color-labeled nucleotides were added, and sequencing was performed with the method of sequencing by synthesis. Image analysis and base-call were performed by the Illumina pipeline, and cDNA sequence tags were revealed after purity filtering. The tags passing initial quality tests were sorted and counted. Each tunnel generates millions of raw reads with sequencing length of 35 bp (target tags plus 3′adaptor). Each molecule in the library represented a single tag derived from a single transcript.
Analysis and mapping of gene expression (DGE) tags
Clean-tags were obtained by filtering the adaptor sequences and removing low-quality sequences (containing ambiguous bases) and then mapped to the reference genome and genes, which were available at ftp://maizesequence.org/pub/maize/release-5b. Only the tags with perfect match or one mismatch were further considered and annotated based on the reference genes. The expression level of each gene was estimated by the frequency of clean tags and then normalized to TPM (number of transcripts per million clean tags) [1], which is a standard method and extensively used in DGE analysis [43]. The expression level of each gene was measured by the normalized number of matched clean tags, and then KOG functional classification, Gene Ontology (GO) and pathway annotation and enrichment analyses were based on the NCBI COG (http://www.ncbi.nlm.nih.gov/COG) [52], Gene Ontology Database (http://www.geneontology.org/) [23] and KEGG pathway (http://www.genome.jp/kegg/)(Ogata et al. [46],respectively.
Identification of different expression genes in two libraries
The probability that one gene G is equally expressed in two samples can be illustrated by the following formula:
N1 and N2 denotes the total number of clean tags in two compared libraries, respectively, while x and y represents the clean tags mapping to gene G. P value indicates the significance of prospect differences of transcript accumulation. A combination of FDR < 0.001 and the absolute value of log2-Ratio >= 1 were used as the threshold to determine the significance of gene expression difference in this research.
GO and pathway enrichment analysis of DEGs
We obtained the GO terms for each maize gene using Blast2GO (version 2.3.5) (http://www.blast2go.org/) with the default parameters. Blast2GO was also used for a GO functional enrichment analysis of certain genes, by performing Fisher’s exact test with a robust FDR correction to obtain an adjusted P value between certain test gene groups and the whole genome annotation.
Validate the DGEs by Real-time PCR
To detect the expression patterns of candidate genes, high-resistance maize inbred line R15 was used in this study, [59] total RNA was isolated form maize leaf sheaths exposed to R. solani with 36, 40, 44, 48 h. Control samples were harvested from water-treated leaf sheaths incubated under the same conditions. To validate the DGEs obtained from Solexa sequencing, 12 DGEs were subjected to quantitative real-time PCR analysis using ABI7500. Actin1 (GRMZM2G126010) was used as the endogenous control and cDNA synthesis was carried out using 1 μg total RNA. The corresponding primers were designed by primer5 software and listed in (Support information Table S2) The amplification programs were performed according to the standard protocol of the ABI7500 system: 95 °C for 30 s; 95 °C for 5 s, 60 °C for 30 s, 40 cycles, and followed by a thermal denaturing step to generate the melt curves for verification of amplification specificity. All reactions were run in triplicate, including non-template controls. The threshold cycles (Ct) of each tested genes were averaged for triplicate reactions and the values were normalized according to the Ct of the control products of Actin1 gene. Statistical analysis was performed using the 2-△△CT method.
Results
Characterization of the sequenced Solexa libraries
To identify genes involved in maize sheaths infected by R. solani, two maize Solexa libraries were constructed from tissues of maize sheaths without any leaves or development leaves, including 4-d and 2-ck libraries. Sequencing depths of 4,200,000 and 4,200,000 tags were achieved in the two libraries respectively, after filtering low quality tags (tags containing ‘N’ and adaptor sequences), 4,006,778 and 3,995,176 tags (distinct tags) were remained in 4-d and 2-ck libraries respectively. Considering of the robustness of subsequently data analysis, tags recorded only once were first wiped off owing to their unreliability; leaving 193,222 and 204,824 distinct tags in each library that were detected multiple times (clean tags). In addition, there were 11,602 more unique tags in the 4-d than in the 2-ck library, possibly representing genes related to pathogen interaction and symptom development. The frequency of these tags is shown in (Table 1), which comprises copy numbers in the range 2–100 or higher, of which, the majority of clean tags (62.9 % from each) were present at low copy numbers (<10 copies), and approximately 33.5 % tags from each library were counted between 11 and 100 times. Only approximately 3.6 % tags were detected more than 100 times. The percentage of unique tags rapidly declined as copy number increased, indicating only a small portion of the transcripts was expressed at high level in the conditions tested.
Annotation of clean tag
To identify the genes corresponding to the 193,222 and 204,824 meaningful tags in each library, the clean tags were mapped to the reference database [25, 37]. In this study, we used blastn to map the unique tags against the reference genome and gene sequences of maize respectively, an essential dataset containing 32,540 reference genes expressed in the maize genome (http://maizesequence.org/index.html) was prepared by expressed gene analysis. Only the clean tags that matched perfectly or with one mismatch were analyzed further. By assigning the experimental Solexa tags to the virtual reference ones (Table 2), we observed that 159,532(56.7 %) and 110,518 (53.9 %) tags were perfectly matched to the reference genes in 2-ck and 4-d respectively. In addition, approximately 15 % tags in two libraries were mapped to the antisense strands, demonstrating that those regions might be bidirectional transcribed. However, the large proportion of non-matched clean tags revealed that the efficiency of annotation was low when the copy number was between two and five, which is in accordance with the studies of the transcriptomes of zebrafish.
It is shown that 4-d and 2-ck libraries were sequenced to saturation in Fig. 1, only fewer tags were identified as the number of sequencing tags increases, producing a full representation of the transcripts in the conditions tested. In conclusion, 4-d library has the relative higher mapping efficiency than 2-CK, indicates that more transcripts have been expressed in development of sheath tissues infected by R. solani in 4-d library than in 2-CK.
Comparison and analysis of differentially expressed genes
After mapping the tags against the reference genes, the count of the tag corresponding to each gene was calculated in each of the two libraries and used to estimate the gene expression level. The transcripts detected with at least two-fold differences (FDR < 0.001 and absolute values of log2 (Ratio) >= 1) in the 4-d compare with the 2-CK are shown in Fig. 2. The statistic difference of accumulation of unique tags between them was shown in Fig. 3. To study a subset of genes that were associated with maize sheath infected by R. solani and to assess the molecular basis of its disease-resistant, we analyzed the most differentially regulated tags with a log2 ratio >2 or <−2 using a greater statistically significant value (P < 0.001) as well as false discovery rates (FDR < 0.01), representing 1,476 up-regulated and 1,754 down-regulated transcripts. According to the Venn diagram, there are 1,610 differential genes in 2-ck and 1,553 in 4-d respectively in Fig. 4. Apart from the unknown transcripts (55 %), predicted or known genes were categorized according to their functions. GO functional annotation of DEGs indicated that both up-regulated genes and down-regulated ones can be classified into 11 categories in Fig. 5, such as catalytic, electrical carrier, transcription regulation and enzyme regulator, and so forth. Significant GO analysis of DEGs in molecular function showed that the top one was transferase activity, transferring hexosyl groups, it is interesting that most of DEGs involved in biological process such as response to stimulus, multicellular organismal process and response to abiotic stimulus in (Support information Table 3). Among these highly expressed genes, some of which were associated with defense, transport, transcription, signal transduction and metabolism and the others were associated with senescence, abiotic and biotic stresses. (Support information Table S1).
Compare gene expression in two libraries. Gene Expression between 2-ck and 4-d (treat by BLSB) was analyzed, respectively. Blue dots represent the transcripts with no significant expression. Red dots and green dots represent transcripts more abundant in the stage sample and control, respectively. “FDR < 0.001” and “absolute value of log2 Ratio ≥ 1” were used as the thresholds to judge the significance of gene expression difference. (Color figure online)
Tags with different expression in stage samples (6-d) compared to control sample (2-ck). Red region represents the differentially expressed tags with differentia expression less than five folds. Blue and green region represent the up- and down-regulated tags for more than five folds, respectively. (Color figure online)
Pathway enrichment analysis of DEGs
The R. sonali affected biological pathways were evaluated by enrichment analysis of DEGs. Significantly enriched metabolic pathways and signal transduction pathways were identified. A total of 96 and 100 pathways were affected by up-regulated DEGs and down-regulated DEGs, respectively (Support information Table 5), in addition to 11 KO numbers with different expression genes between 4-d and 2-ck were not contained in KEGG (Support information Table 6), most of them were found in the enriched pathways and the first twenty enriched pathways were reported in (Support information Table 4). Pathways with Q value <0.05 are considerably enriched. Caffeine metabolism proteins constituted the only significantly affected pathway for the upregulated DEGs (Q < 0.05). Other non-significant enriched pathways with a large number of unregulated DEGs included betalain biosynthesis and Nicotinate and nicotinamide, ether lipid metabolism, non-homologous end-joining and proteasome. Moreover, there were more significantly enriched pathways for the down-regulated DEGs, which were involved in photosynthesis, as well as metabolism of vitamin B6, sphingolipid, arachidonic acid and degradation of glycosaminoglycan, along with biosynthesis of N-glycan, carotenoid and fatty acid elongation in mitochondria.
Quantitative real-time PCR (qRT-PCR) confirmation
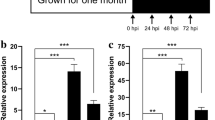
To evaluate the validity the DGEs obtained from Solexa sequencing and to further assess the patterns of differential gene expression, 12 candidate genes were selected and detected by qRT-PCR, including 8 up-regulated DGEs and 4 down-regulated DGEs (Support information Table S2). As shown in Fig. 6, the expression patterns showed general agreement with the Solexa sequencing, but the apparent discrepancies with respect to ratio was clearly found, transcripts from highly abundant Solexa tags appeared at the expected lower cycle numbers in the quantitative PCR analyses. It should be attributed to the essentially different algorithms determined by the two techniques (DEG and Q-PCR).
The 12 candidate DGEs validated by Q-PCR technology. Note: A-L represented the DGEs were as follows respectively. A: GRMZM2G397755, B: GRMZM2G151425, C: GRMZM2G332390, D: GRMZM2G012242, E: GRMZM2G113860, F: GRMZM2G136372, G: GRMZM2G123119, H: GRMZM2G179896, I: GRMZM2G065203, J: GRMZM2G085924,K: GRMZM2G059191, L: GRMZM2G303419.Their function and expression levels in DGE library were shown in Table S2
Discussion
The major goal of the present study is to preliminarily explore disease-resistant transcripts involved in the maize sheath infected by R. solani, as well as to provide foundation for investigating their regulation mechanisms. To our knowledge, this is the first report that comprehensively shows the transcriptional changes during the development of maize sheath inoculated with R. solani. We used the Illumina/Solexa DGE system, which is essentially a serial analysis of gene expression-based tag profiling approach. The results provided estimates of gene expression as determined by the frequency that any given tag (representing a transcript) is sequenced (Fig. 2). The data indicates that there is sufficient coverage depth to reach saturation, that is, a complete assessment of all transcripts present in the libraries. In addition, expression levels of candidate genes were confirmed by Q-PCR technology. Although the differences in gene expression did not match the magnitude of those detected by Solexa-based sequencing method, the trends of up- and down- regulation were similar, it may be owe to the apparent discrepancies with respect to ratio and the essentially different algorithms determined by the two techniques. In conclusion, Solexa sequencing has been documented to be more sensitive for estimation of gene expression, especially for low-abundance transcripts compared to microarrays and Real-time RT-PCR [1].
Functional analysis of DEGs
A set of transcripts was clearly more abundant in leaf sheaths after R. solani infection in 4-d compared to 2-CK. This group possibly contains elements that confer resistance to the spread of the pathogen in “R15”. These transcripts likely encode genes responding to the pathogen that underlie genetic resistance, which were broadly grouped into the following categories based on their known roles combing GO enrichment analysis with scientific literature.
Defense response genes
Among defense response genes, in addition to thaumatin-like protein [47], plant disease resistance response protein, Flagellar hook-length control protein, and harpin-induced protein-related [16, 28] were found in our research, major facilitator superfamily, Mlo-related protein, pathogenesis-related transcriptional factor, pectinesterase inhibitor and pistil-specific extensin-like protein were also enriched in this study which have been widely studied in plant pathogen resistance. Moreover, a new transcript, universal stress protein A was found in this study, which has been best characterized and found to be highly expressed in response to heat, substrate starvation, exposure to antimicrobial agents and oxidative stress in Escherichia coli K-12. In our research, Universal stress protein A (+11.03 fold in 4-d vs 2-ck) showed the great trend in 4-d than 2-CK and was found associated with R. solani attack and may also play an important role in respond to R. solani attack.
Transport
Several transcripts associated with transport function were found in this research. Multi antimicrobial extrusion protein MatE (+4.58 fold in 4-d vs 2-d) and ABC transporter (+5.58 fold in 4-d vs 2-CK) are well known transporters in clinical study for bacterial infection of human [41]. In plant, plant ABC transporters have been demonstrated to participate in chlorophyll biosynthesis, formation of Fe/S clusters, stomatal movement, and probably ion fluxes; hence they may play a central role in plant growth and developmental processes. And plant mates have been genetically identified, characterized and found to be involved in the detoxification of endogenous secondary metabolites and xenobiotics. Another gene identified to be transport related is mitochondrial carrier protein (+11.17 fold in 4-d vs 2-CK) which might be involved in the excretion of organic acids and rhizotoxic aluminum tolerance [15]. Moreover, sugar transporter and ATPase, P-type, K/Mg/Cd/Cu/Zn/Na/Ca/Na/H-transporter were also demonstrated in our study, however, there was only a little report about these transporters, which might play a purely nutritional role and supply sugars and K/Mg/Cd/Cu/Zn/Na/Ca/Na/H to cells for growth and development, whereas others are involved in generating osmotic gradients required to drive mass flow or movement.
Signal transduction
There were 36 transcripts in our results associated with signal transduction. Four came from genes (GRMZM2G048131, GRMZM2G074742, GRMZM2G121309, GRMZM2G163848) encoding AUX/IAA proteins that were more prevalent (4.5–6.2 fold) in the 4-d library than in 2-CK. It is reported that exogenous salicylic acid (SA) treatment triggers the stabilization of AUX-IAA (auxin/indole-3-acetic acid) proteins that negatively regulate auxin signaling, thus attenuating auxin signaling [58], probably indirectly through repression of the F-box auxin receptor TIR1, thereby decreasing sensitivity to auxin treatment and limiting ubiquitination of AXR2 and other AUX-IAA proteins. Various signals presented in our results, including auxin, abscisic acid (ABA), as well as intracellular messengers like calcium has been proposed to regulate plant responses in adverse environmental conditions and thus contribute to the coordination of plant stress physiology [63]. Activation of ABA biosynthetic and signaling pathways promotes disease susceptibility to several plant pathogens [5]; Ton and Mauch-Mani [53], and most of RING-H2 genes were annotated in this study which has demonstrated regulatory function in ABA signaling, drought tolerance, regulation of growth and defense responses against abiotic/biotic stresses [30, 38]. Moreover, six transcripts (GRMZM2G041729, GRMZM2G419452, GRMZM2G040094, GRMZM2G366411, GRMZM2G001814, and GRMZM2G021998) were associated with calcium signaling pathway. All of these are also induced by senescence and stresses [18, 34].
Transcription
49 transcripts associated with tyrosine protein kinase transcription were 4–8.033 fold more abundant in 4-d than 2-CK libraries. Transcripts annotated as zinc-finger protein 1, Transcriptional factor B3, AP2 domain class transcription factor, basic helix-loop-helix protein, DNA-binding WRKY, Calcium-binding EF-hand, Leucine-rich repeat protein were all present at higher steady state levels in infected tissue. They have been documented to play important roles in responding to phytohormone stasis, pathogen attack and environmental stresses [17, 19, 20, 51] and all of which have been observed to increase in response to pathogen challenge [50]. And also these defense-associated TFs can regulate downstream defense-related genes, and may be regulated by phosphorylation themselves [31].
Metabolism
Protein metabolism
Several transcripts related to protein metabolism were found abundant in 4-d library, Among them, protein phosphatase 2c (4.58–5.58 fold in 4-d VS 2-CK) regulates numerous ABA responses [22, 57]. Recent studies place protein phosphatases in various signaling cascades including those for ABA, pathogen and stress responses, and developmental processes. It is clearly demonstrated that protein phosphatases function not only by counterbalancing the protein kinases but also by taking a leading role in many signaling events [40]. Protein kinase (4.58–6.57 fold in 4-d VS 2-CK) play a central role in signaling during pathogen recognition and the subsequent activation of plant defence mechanisms [48]. In addition, phosphate-induced protein 1(GRMZM2G119766) has been shown to be involved in MAP kinase pathways. In plants, there is evidence for MAPKs playing a role in the signaling of abiotic stresses, pathogens and plant hormones.
Secondary metabolism
This subcategory contained four genes, including a higher level of polyketide synthase, (4.48–5.58 fold in 4-d vs CK) in infected tissues, it is consistent with previous reports that it will be more abundant after pathogen infection and the secondary metabolite synthesis depends on polyketide synthase[6]. Isopenicillin N synthase (GRMZM2G031432, GRMZM2G0317241, 5.58 and 6.89 fold in 4-d vs CK) has also played a potential pathogen resistance role because it is involved in biosynthesis of Peroxisomes, an important product in resistance to pathogen infection[6]. UDP-glucose glucosyltransferase (+ 4.28–7.82 fold in 4-d vs 2-CK) are reported to be induced by abiotic stresses [21, 45].
In addition, other highly expressed metabolic genes in the 4-d samples were plant lipid transfer protein, cytochrome P450 and thioredoxin-like. It is reported that the role of LTP in the physiology of plants, including plant defense against phytopathogens [9, 55, 65]. Cytochrome P450 and thioredoxin-like are related to carbohydrate metabolism, photosynthesis and thioredoxin synthesis. Thioredoxins are ubiquitous disulfide reductases that regulate the redox status of target proteins which was involved in oxidative damage avoidance by supplying reducing power to reductases detoxifying lipid hydroperoxides or repairing oxidized proteins and could act as regulators of scavenging mechanisms and as components of signalling pathways in the plant antioxidant network [56].
Transcripts less abundant in infected sheaths
The most striking functions for transcripts less abundant in infected sheath were those associated with carotenoid biosynthesis. DEGs were detected to be less prevalent in the 4-d libraries more than fourfold compared to CK, most of which, such as O-methyltransferase, COMT [24], ferredoxin–NADP reductase [10], phospholipid/glycerol acyltransferase [11], S-adenosyl-l-Met (SAM) dependent carboxyl methyltransferase [33].Of which, O-methyltransferase, COMT is reported to be positively associated with plant defense responses to pathogen attack. However, our data indicated that the expression level of these transcripts was lower in infected sheath (Fig. 6).
Ferredoxin-NADP+(oxido)reductase (EC 1.18.1.2, FNR) is an FAD-containing enzyme that catalyzes the reversible electron transfer between NADP(H) and electron carrier proteins such as ferredoxin and flavodoxin [3]. Of which, ferredoxin (Fd) is a fundamental protein that is involved in several metabolic pathways. The amount of Fd found in plants is generally regulated by environmental stress, including biotic and abiotic events and Fd levels were increased by inoculation with Pseudomonas syringae pv. syringae but were reduced by Erwinia carotovora ssp. Carotovora [26], but we found Fd levels were reduced by BLSB through reducing Ferredoxin-NADP+(oxido)reductase in this research. In addition, it is reported that the acyl-CoA-independent synthesis of TAG is mediated by phospholipid:diacylglycerol acyltransferase in plants and it is proposed to be involved in the accumulation of high levels of hydroxylated fatty acid (ricinoleic acid) and epoxidated fatty acid (vernolic acid) [14], these phospholipid-derived molecules are emerging as novel second messengers in plant defence signalling. They may be regulate ion channels and proton pumps by free fatty acids, and then conversion free fatty acids into bioactive octadecanoids such as jasmonic acid to response to pathogen attack. Moreover, SAM dependent carboxyl methyltransferase, the first characterized member of the family, catalyzes the formation of methyl salicylate (MeSA) from SA and SAM [44, 49], and MeSA is component of the floral scents of some plants and are also produced by vegetative parts of plants in response to environmental challenge [12].
Pathway enrichment analysis of DEGs
Pathway enrichment analysis revealed the most significantly affected pathways during the R. solani infection in“R15”. It is not surprising that the “metabolic pathways” was the most affected for the DEGs in 4-d library. This finding implies that the maize resistance material R15 utilizes biosynthesis of plant hormones, terpenoids and steroids, N-glycan, Zeatin to protect itself from the pathogen attack. The second affected pathway was the “Plant-pathogen interaction” pathway. In this pathway genes encoding WRKY family were more prevalent in the 4-d than the 2-ck library. In addition, genes required for plant-pathogen interaction were also affected, such as CDPK, NOS, and JAZ. These genes are involved in the plant-pathogen interaction system, and may regulate plant defense patterns in response to pathogen, thereby linking signaling with metabolism. The other noticeable pathways with a large amount of DEGs associated with BLSB infection were starch and sucrose metabolism, Ubiquitin mediated proteolysis, plant hormone biosynthesis, and splicesome-associated proteins. For DEGs less prevalent in infected versus control libraries, there was significant enrichment for transcripts associated with photosynthesis.
Conclusions
Solexa-based sequencing can be used for analyzing variation in gene expression between two samples. The gene expression level in high-resistance maize inbred line “R15” sheath infection with R. solani (4-d) changed significantly in comparison with control (2-ck). Analysis of differential expressed genes involved in response to pathogen infection allows delineation of candidate genes potentially relevant to R. solani resistance in maize. In addition, Our analysis of the transcription and gene expression in “R15” infection by R. solani revealed changes in multiple signaling pathways involved in immunity in “R15”, the candidate immune-related genes and association signaling pathways involved in pathogen infection were identified and thereby provided valuable leads for further investigation the immune response in maize.
References
AC’t Hoen P, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RHAM, De Menezes RX, Boer JM, Van Ommen GJB, Den Dunnen JT (2008) Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res 36:e141–e141
Alexander D, Goodman RM, Gut-Rella M, Glascock C, Weymann K, Friedrich L, Maddox D, Ahl-Goy P, Luntz T, Ward E (1993) Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc Natl Acad Sci 90:7327
Arakaki A, Ceccarelli EA, Carrillo N (1997) Plant-type ferredoxin-NADP+reductases: a basal structural framework and a multiplicity of functions. FASEB J 11:133–140
Asmann YW, Klee EW, Thompson EA, Perez EA, Middha S, Oberg AL, Therneau TM, Smith DI, Poland GA, Wieben ED (2009) 3′tag digital gene expression profiling of human brain and universal reference RNA using Illumina Genome Analyzer. BMC Genom 10:531
Asselbergh B, Curvers K, França SC, Audenaert K, Vuylsteke M, Van Breusegem F, Höfte M (2007) Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol 144:1863–1877
Böhnert HU, Fudal I, Dioh W, Tharreau D, Notteghem JL, Lebrun MH (2004) A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16:2499–2513
Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18:630–634
Campbell MA, Fitzgerald HA, Ronald PC (2002) Engineering pathogen resistance in crop plants. Transgenic Res 11:599–613
Carvalho AO, Gomes VM (2007) Role of plant lipid transfer proteins in plant cell physiology-a concise review. Peptides 28:1144–1153
Chaki M, Valderrama R, Fernández-Ocaña ANAM, Carreras A, Gómez-Rodríguez MV, López-Jaramillo J, Begara-Morales JC, Sánchez-Calvo B, Luque F, Leterrier M (2011) High temperature triggers the metabolism of S-nitrosothiols in sunflower mediating a process of nitrosative stress which provokes the inhibition of ferredoxin–NADP reductase by tyrosine nitration. Plant Cell Environ 34:1803–1818
Chanda B, Venugopal SC, Kulshrestha S, Navarre DA, Downie B, Vaillancourt L, Kachroo A, Kachroo P (2008) Glycerol-3-phosphate levels are associated with basal resistance to the hemibiotrophic fungus Colletotrichum higginsianum in Arabidopsis. Plant Physiol 147:2017–2029
Chen F, D’Auria JC, Tholl D, Ross JR, Gershenzon J, Noel JP, Pichersky E (2003) An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J 36:577–588
Chen J, Tang C, Gao Z, XUE C, NIU X, SONG Z (2000) On penetration process of sheath blight pathogen in maize. J Shenyang Agric Univ 31:503–506
Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S (2000) Phospholipid: diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci 97:6487
Deng W, Luo K, Li Z, Yang Y (2008) Molecular cloning and characterization of a mitochondrial dicarboxylate/tricarboxylate transporter gene in Citrus junos response to aluminum stress. Mitochondrial DNA 19:376–384
Ditt RF, Kerr KF, De Figueiredo P, Delrow J, Comai L, Nester EW (2006) The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol Plant Microbe Interact 19:665–681
Duek PD, Fankhauser C (2005) bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci 10:51–54
Espinoza C, Medina C, Somerville S, Arce-Johnson P (2007) Senescence-associated genes induced during compatible viral interactions with grapevine and Arabidopsis. J Exp Bot 58:3197–3212
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–205
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371
Gachon CMM, Langlois-Meurinne M, Saindrenan P (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10:542–549
Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11:1897–1910
Harris M, Deegan J, Lomax J, Ashburner M, Tweedie S, Carbon S, Lewis S, Mungall C, Day-Richter J, Eilbeck K (2008) The gene ontology project in 2008. Nucleic Acids Res 36:D440–D444
He J, Chen F, Chen S, Guan Z, Fang W, Liu Z, Lu J (2010). Expression Profiles of Defense-Related Genes in Response to Aphid Inoculation in Different Chrysanthemum Cultivars. XXVIII International Horticultural Congress on Science and Horticulture for People (IHC2010): International Symposium on 929. pp. 51-57)
Hegedűs Z, Zakrzewska A, Ágoston VC, Ordas A, Rácz P, Mink M, Spaink HP, Meijer AH (2009) Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. Mol Immunol 46:2918–2930
Huang H-E, Ger M-J, Chen C-Y, Pandey A-K, Yip M-K, Chou H-W, Feng T-Y (2007) Disease resistance to bacterial pathogens affected by the amount of ferredoxin-I protein in plants. Mol Plant Pathol 8:129–137
Huang M, Tan J, Yang J, Yang K (2007) Research advance on Banded Leaf and Sheath Blight of maize [J]. Southwest China J Agric Sci 2:006
Jolivet K, Grenier E, Bouchet JP, Esquibet M, Kerlan MC, Caromel B, Mugniery D, Lefebvre V (2007) Identification of plant genes regulated in resistant potato Solanum sparsipilum during the early stages of infection by Globodera pallida. Genome 50:422–427
Jongeneel CV, Iseli C, Stevenson BJ, Riggins GJ, Lal A, Mackay A, Harris RA, O’Hare MJ, Neville AM, Simpson AJG (2003) Comprehensive sampling of gene expression in human cell lines with massively parallel signature sequencing. Proc Natl Acad Sci 100:4702–4705
Ko JH, Yang SH, Han KH (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47:343–355
Korfhage U, Trezzini GF, Meier I, Hahlbrock K, Somssich IE (1994) Plant homeodomain protein involved in transcriptional regulation of a pathogen defense-related gene. Plant Cell 6:695–708
Krishnaveni S, Muthukrishnan S, Liang G, Wilde G, Manickam A (1999) Induction of chitinases and β-1, 3-glucanases in resistant and susceptible cultivars of sorghum in response to insect attack, fungal infection and wounding. Plant Sci 144:9–16
Kwon S, Hamada K, Matsuyama A, Yasuda M, Nakashita H, Yamakawa T (2009) Biotic and abiotic stresses induce AbSAMT1, encoding S-adenosyl-l-methionine: salicylic acid carboxyl methyltransferase, in Atropa belladonna. Plant Biotechnol 26:207–215
Li A, Wang X, Leseberg CH, Jia J, Mao L (2008) Biotic and abiotic stress responses through calcium-dependent protein kinase (CDPK) signaling in wheat (Triticum aestivum L.). Plant Signal Behav 3:654
Li L, YongYi M, ZhiMing Z, GuangTang P, MaoJun Z (2009) cDNA-AFLP analysis of Rhizoctonia solani AG1-IA-induced maize differential gene expression. Acta Phytopathol Sin 39:385–391
Li L, YongYi M, ZhiMing Z, PengFei L, GuangTang P, MaoJun Z (2009) Functional effects of different defense enzymes on banded leaf and sheath blight of maize. Journal of Maize Sciences 17:99–102, 106
Li YJ, Fu YR, Huang JG, Wu CA, Zheng CC (2011) Transcript profiling during the early development of the maize brace root via Solexa sequencing. FEBS J 278:156–166
Liu H, Zhang H, Yang Y, Li G, Wang X, Basnayake BMVS, Li D, Song F (2008) Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses. Plant Mol Biol 68:17–30
Liu L, Zhang Z, Zhao M, Wang J, Lin H, Shen Y, Pan G (2011) Molecular cloning and characterization of pathogenesis-related protein 5 in Zea mays and its antifungal activity against Rhizoctonia solani. Afr J Biotechnol 10:19286–19293
Luan S (1998) Protein phosphatases and signaling cascades in higher plants. Trends Plant Sci 3:271–275
Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kolukisaoglu U, Müller-Röber B, Schulz B (2002) Multifunctionality of plant ABC transporters-more than just detoxifiers. Planta 214:345–355
Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S (2004) The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res 14:1641–1653
Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, Jones S, Zhao Y, Hirst M, Marra MA (2009) Next-generation tag sequencing for cancer gene expression profiling. Genome Res 19:1825–1835
Negre F, Kolosova N, Knoll J, Kish CM, Dudareva N (2002) Novel S-adenosyl–methionine: salicylic acid carboxyl methyltransferase, an enzyme responsible for biosynthesis of methyl salicylate and methyl benzoate, is not involved in floral scent production in snapdragon flowers. Arch Biochem Biophys 406:261–270
Nishizawa-Yokoi A, Yabuta Y, Shigeoka S (2008) The contribution of carbohydrates including raffinose family oligosaccharides and sugar alcohols to protection of plant cells from oxidative damage. Plant Signal Behav 3:1016
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27:29–34
Punja ZK (2001) Genetic engineering of plants to enhance resistance to fungal pathogens–a review of progress and future prospects. Can J Plant Pathol 23:216–235
Romeis T (2001) Protein kinases in the plant defence response. Curr Opin Plant Biol 4:407–414
Ross JR, Nam KH, D’Auria JC, Pichersky E (1999) S-Adenosyl–Methionine: Salicylic Acid Carboxyl Methyltransferase, an Enzyme Involved in Floral Scent Production and Plant Defense, Represents a New Class of Plant Methyltransferases. Arch Biochem Biophys 367:9–16
Singh KB, Foley RC, Oñate-Sánchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5:430–436
Takatsuji H (1998) Zinc-finger transcription factors in plants. Cell Mol Life Sci 54:582–596
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41
Ton J, Mauch-Mani B (2004) Β-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J 38:119–130
Tsror L (2010) Biology, epidemiology and management of Rhizoctonia solani on potato. J Phytopathol 158:649–658
Velazhahan R, Radhajeyalakshmi R, Thangavelu R, Muthukrishnan S (2001) An antifungal protein purified from pearl millet seeds shows sequence homology to lipid transfer proteins. Biol Plant 44:417–421
Vieira Dos Santos C, Rey P (2006) Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci 11:329–334
Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurièrea C, Merlot S (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21:3170–3184
Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X (2007) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17:1784–1790
Yang A, Pan G, Ye H, Tang L, Rong T (2003) Evaluating resistance of inbred lines of corn to maize sheath blight and screening of resistance resources. Plant Prot Beijing 29:25–27
Zala M, McDonald B, DE ASSIS JB, Ciampi M, Storari M, Peyer P, Ceresini P (2008) Highly polymorphic microsatellite loci in the rice- and maize-infecting fungal pathogen Rhizoctonia solani anastomosis group 1 IA. Mol Ecol Resour 8:686–689
Zhang Z, Li W, Zhao M, Rong T, Pan G, Jeffers DP, Zhang S (2006) Quantitative trait loci for resistance to banded leaf and sheath blight in maize. Crop Sci 46:1039–1045
Zhang Z, Liu L, Lin H, Yuan G, Zeng X, Shen Y, Zhao M, Zhao Q, Pan G (2012) Identification of genes differentially expressed in maize (Zea mays L.) during Rhizoctonia Solani Kühn infection by suppression subtractive hybridization. Afr J Biotechnol 11:2827–2838
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247
Zhu T, Song F, Zheng Z (2006) Molecular characterization of the rice pathogenesis-related protein, OsPR-4b, and its antifungal activity against Rhizoctonia solani. J Phytopathol 154:378–384
Zottich U, Da Cunha M, Carvalho AO, Dias GB, Silva N, Santos IS, do Nacimento VV, Miguel EC, Machado OLT, Gomes VM (2011) Purification, biochemical characterization and antifungal activity of a new lipid transfer protein (LTP) from Coffea canephora seeds with α-amylase inhibitor properties. Biochim Biophys Acta 1810:375–383
Acknowledgments
This work was supported by the grants from the National Natural Science Foundation of China (30900901), the Research Project on Rice Functional Genes related to the Maize Large-scale Gene Discovery and Functional Genomics Resource, Technology, Information Platform Construction Program (863) of China (SS2012AA100107, 2012AA10A300), Ph.D. Programs Foundation of Ministry of Education of China (20095103120002) and the Major Project of China on New varieties of GMO Cultivation (2011ZX08003-003) and the Youth Natural Science Foundation of Department of Education of Sichuan Province of China (11ZB123).
Author information
Authors and Affiliations
Corresponding author
Additional information
Jian Gao and Zhe Che have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Gao, J., Chen, Z., Luo, M. et al. Genome expression profile analysis of the maize sheath in response to inoculation to R. solani . Mol Biol Rep 41, 2471–2483 (2014). https://doi.org/10.1007/s11033-014-3103-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3103-z