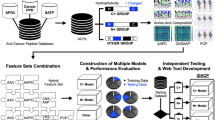
Abstract
Conventional cancer therapies are highly expensive and have serious complications. An alternative approach now emphasizes on the development of small, biologically active peptides without acute toxicity. Experimental screening to find curative anticancer peptides (ACP) often gives rise to multiple obstacles and is time dependent. Consequently, developing an effective computational technique to identify promising ACP candidates prior to preclinical research is in high demand. This study proposed a machine-learning framework that used the light gradient-boosting machine as a classifier and two compositional and two binary profile features as input. The ensemble model displayed an accuracy, MCC, and AUROC of 97.52%, 0.91, and 0.98, respectively, which outclassed most of the existing sequence-based computational tools. A distinct dataset of non-mutagenic, non-toxic, and non-inhibitory Cytochrome P-450 peptides was used to validate the hybrid model. The most relevant ACP in the alternative dataset was compared with two standard ACPs, beta defensin 2, and cecropin-A. Molecular docking of the predicted peptide revealed that it has a strong binding affinity with twenty-five anticancer drug targets, most notably phosphoenolpyruvate carboxykinase (− 7.2 kcal/mol). Additionally, molecular dynamics simulation and principal component analysis supported the stability of the peptide-receptor complex. Overall, the present findings will take a step forward in rational drug design through rapid identification and screening of therapeutic peptides.






Similar content being viewed by others
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA Cancer J Clin 72:7–33. https://doi.org/10.3322/caac.21708
Fitzgerald RC, Antoniou AC, Fruk L, Rosenfeld N (2022) The future of early cancer detection. Nat Med 28:666–677. https://doi.org/10.1038/s41591-022-01746-x
Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B (2017) The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull 7:339–348. https://doi.org/10.15171/apb.2017.041
Zahreddine H, Borden KLB (2013) Mechanisms and insights into drug resistance in cancer. Front Pharmacol 14:4–28. https://doi.org/10.3389/fphar.2013.00028
Lin MW, Tseng YW, Shen CC, Hsu MN et al (2018) Synthetic switch-based baculovirus for transgene expression control and selective killing of hepatocellular carcinoma cells. Nucleic Acids Res 46(15):e93. https://doi.org/10.1093/nar/gky447
de Souza JA, Wong YN (2013) Financial distress in cancer patients. J Med Person. https://doi.org/10.1007/s12682-013-0152-3
Omenn GS (2016) Strategies for genomic and proteomic profiling of cancers. Stat Biosci 8:1–7. https://doi.org/10.1007/s12561-014-9111-7
Basith S, Manavalan B, Shin TH, Lee G (2020) Machine intelligence in peptide therapeutics: a next-generation tool for rapid disease screening. Med Res Rev 40:1276–1314. https://doi.org/10.1002/med.21658
Charoenkwan P, Chiangjong W, Lee VS, Nantasenamat C et al (2021) Improved prediction and characterization of anticancer activities of peptides using a novel flexible scoring card method. Sci Rep 11(1):3017. https://doi.org/10.1038/s41598-021-82513-9
Chen J, Cheong HH, Siu SWI (2021) xDeep-AcPEP: deep learning method for anticancer peptide activity prediction based on convolutional neural network and multitask learning. J Chem Info Model 61(8):3789–3803. https://doi.org/10.1021/acs.jcim.1c00181
Tornesello AL, Borrelli A, Buonaguro L, Buonaguro FM, Tornesello ML (2020) Antimicrobial peptides as anticancer agents: functional properties and biological activities. Molecules 19(12):2850. https://doi.org/10.3390/molecules25122850
Karpiński TM, Adamczak A (2018) Anticancer activity of bacterial proteins and peptides. Pharmaceutics 10:10020054. https://doi.org/10.3390/pharmaceutics10020054
Kondo E, Iioka H, Saito K (2021) Tumor-homing peptide and its utility for advanced cancer medicine. Cancer Sci 112:2118–2125. https://doi.org/10.1111/cas.14909
Wang L, Wang N, Zhang W, Cheng X et al (2022) Therapeutic peptides: current applications and future directions. Sig Transduct Target Ther 7(1):48. https://doi.org/10.1038/s41392-022-00904-4
Usmani SS, Bedi G, Samuel JS, Singh S et al (2017) THPdb: database of FDA-approved peptide and protein therapeutics. PLoS ONE 12(7):e0181748. https://doi.org/10.1371/journal.pone.0181748
Muttenthaler M, King GF, Adams DJ, Alewood PF (2021) Trends in peptide drug discovery. Nat Rev Drug Discov 20:309–325. https://doi.org/10.1038/s41573-020-00135-8
Chen XG, Zhang W, Yang X, Li C, Chen H (2021) ACP-DA: improving the prediction of anticancer peptides using data augmentation. Front Genet 12:698477. https://doi.org/10.3389/fgene.2021.698477
Hwang JS, Kim SG, Shin TH, Jang YE, Kwon DH, Lee G (2022) Development of anticancer peptides using artificial intelligence and combinational therapy for cancer therapeutics. Pharmaceutics 14(5):997. https://doi.org/10.3390/pharmaceutics14050997
Nasiri F, Atanaki FF, Behrouzi S, Kavousi K, Bagheri M (2021) CpACpP: in silico cell-penetrating anticancer peptide prediction using a novel bioinformatics framework. ACS Omega 6(30):19846–19859. https://doi.org/10.1021/acsomega.1c02569
Tyagi A, Kapoor P, Kumar R, Chaudhary K et al (2013) In silico models for designing and discovering novel anticancer peptides. Sci Rep 3:1–8. https://doi.org/10.1038/srep02984
Agrawal P, Bhagat D, Mahalwal M, Sharma N et al (2020) AntiCP 2.0: an updated model for predicting anticancer peptides. Brief Bioinform 22(3):bbaa153. https://doi.org/10.1093/bib/bbaa153
Chen W, Ding H, Feng P, Lin H, Chou KC (2021) iACP: a sequence-based tool for identifying anticancer peptides. Oncotarget 7(13):16895. https://doi.org/10.18632/oncotarget.7815
Vijayakumar S, Ptv L (2017) ACPP: a web server for prediction and design of anti-cancer peptides. Int J Pept Res Ther 21:99–106. https://doi.org/10.1007/s10989-014-9435-7
Kabir M, Arif M, Ahmad S, Ali Z et al (2018) Intelligent computational method for discrimination of anticancer peptides by incorporating sequential and evolutionary profiles information. Chemom Intell Lab Syst 182:158–165. https://doi.org/10.1016/j.chemolab.2018.09.007
Schaduangrat N, Nantasenamat C, Prachayasittikul V (1973) Shoombuatong W (2019) ACPred: a computational tool for the prediction and analysis of anticancer peptides. Molecules 24:10. https://doi.org/10.3390/molecules24101973
Wei L, Zhou C, Chen H, Song J, Su R (2018) ACPred-FL: a sequence-based predictor using effective feature representation to improve the prediction of anti-cancer peptides. Bioinformatics 34:4007–4016. https://doi.org/10.1093/bioinformatics/bty451
Manavalan B, Basith S, Shin TH, Choi S et al (2017) MLACP: machine-learning-based prediction of anticancer peptides. Oncotarget 8:77121–77136. https://doi.org/10.18632/oncotarget.20365
Wu C, Gao R, Zhang Y, Marinis YD (2019) PTPD: predicting therapeutic peptides by deep learning and word2vec. BMC Bioinform 20(1):456. https://doi.org/10.1186/s12859-019-3006-z
Boopathi V, Subramaniyam S, Malik A, Lee G, Manavalan B, Yang DC (2019) mACPpred: a support vector machine-based meta-predictor for identification of anticancer peptides. Int J Mol Sci 20(8):1964. https://doi.org/10.3390/ijms20081964
Wei L, Zhou C, Su R, Zou Q (2019) PEPred-Suite: improved and robust prediction of therapeutic peptides using adaptive feature representation learning. Bioinformatics 35:4272–4280. https://doi.org/10.1093/bioinformatics/btz246
Wang H, Zhao J, Zhao H, Li H, Wang J (2021) CL-ACP: a parallel combination of CNN and LSTM anticancer peptide recognition model. BMC Bioinform 22(1):512. https://doi.org/10.1186/s12859-021-04433-9
Wu X, Zeng W, Lin F, Xu P, Li X (2022) Anticancer Peptide prediction via multi-kernel cnn and attention model. Front Genet 13:887894. https://doi.org/10.3389/fgene.2022.887894
Feng G, Yao H, Li C, Liu R et al (2022) ME-ACP: multi-view neural networks with ensemble model for identification of anticancer peptides. Comput Biol Med 145:10549. https://doi.org/10.1101/2021.11.22.469543
Lv Z, Cui F, Zou Q, Zhang L, Xu L (2021) Anticancer peptides prediction with deep representation learning features. Brief Bioinform 22(5):bbab008. https://doi.org/10.1093/bib/bbab008
Yi HC, You ZH, Zhou X, Cheng L et al (2019) ACP-DL: a deep learning long short-term memory model to predict anticancer peptides using high-efficiency feature representation. Mol Ther Nucleic Acids 17:1–9. https://doi.org/10.1016/j.omtn.2019.04.025
Zhao Y, Wang S, Fei W, Feng Y et al (2021) Prediction of anticancer peptides with high efficacy and low toxicity by hybrid model based on 3D structure of peptides. Int J Mol Sci 22(11):5630. https://doi.org/10.3390/ijms22115630
Gurung AB, Ali MA, Lee J, Farah MA, Al-Anazi KM (2021) Molecular docking and dynamics simulation study of bioactive compounds from Ficus carica L. with important anticancer drug targets. PLoS ONE 16(7):e0254035. https://doi.org/10.1371/journal.pone.0254035
Cui W, Aouidate A, Wang S, Yu Q et al (2020) Discovering anti-cancer drugs via computational methods. Front Pharmacol 11:00733. https://doi.org/10.3389/fphar.2020.00733
Timmons PB, Hewage CM (2021) ENNAACT is a novel tool which employs neural networks for anticancer activity classification for therapeutic peptides. Biomed Pharmacother 133:111051. https://doi.org/10.1016/j.biopha.2020.111051
Wang CKL, Kaas Q, Chiche L, Craik DJ (2008) CyBase: a database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res 36:D206–D210. https://doi.org/10.1093/nar/gkm953
Thomas S, Karnik S, Barai RS, Jayaraman VK et al (2010) CAMP: a useful resource for research on antimicrobial peptides. Nucleic Acids Res 38:D774–D780. https://doi.org/10.1093/nar/gkp1021
Wang J, Yin T, Xiao X, He D (2018) StraPep: a structure database of bioactive peptides. Database (Oxford) 2018:bay038. https://doi.org/10.1093/database/bay038
Cheng F, Li W, Zhou Y, Shen J et al (2012) AdmetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 52:3099–3105. https://doi.org/10.1021/ci300367a
Berman HM, Westbrook J, Feng Z, Gilliland G et al (2000) The protein data bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Jumper J, Evans R, Pritzel A, Green T et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Chen F, Wang Z, Wang C, Xu Q et al (2017) Application of reverse docking for target prediction of marine compounds with anti-tumor activity. J Mol Gr Model 77:372–377. https://doi.org/10.1016/j.jmgm.2017.09.015
Kassambara A (2017) Multivariate analysis II: Practical guide to principal component methods. In: R: PCA, M (CA), FAMD, MFA, HCPC, factoextra. vol. 2, STHDA. http://library.lol/main/16BD5874049EA93F7866E5436D7E87B8
Silverman RB, Holladay MW (2014) The Organic chemistry of drug design and drug action, Chapter 1-Introduction. In: Silverman RB, Holladay MW (eds) The organic chemistry of drug design and drug action. Academic Press, Cambridge, pp 1–17. https://doi.org/10.1016/B978-0-12-382030-3.00001-5
Pantsar T, Poso A (2018) Binding affinity via docking: fact and fiction. Molecules 23:23081899. https://doi.org/10.3390/molecules23081899
Yu S, Meng S, Xiang M, Ma H (2021) Phosphoenolpyruvate carboxykinase in cell metabolism: roles and mechanisms beyond gluconeogenesis. Mol Metab 53:101257. https://doi.org/10.1016/j.molmet.2021.101257
Aier I, Varadwaj P, Raj U (2016) Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci Rep 6:34984. https://doi.org/10.1038/srep34984
Funding
Departmental facilities and M. Tech Project funding of NIT Agartala.
Author information
Authors and Affiliations
Contributions
SG received the Bachelor’s degree in Computer Science and Engineering from the Maulana Abul Kalam Azad University of Technology, India. He is presently pursuing the M. Tech programme in Bioengineering department at National Institute of Technology Agartala, India. His research interests include bioinformatics and machine learning. JT received the B. Tech degree from the SRM Institute of Technology, India. She is currently enrolled in the M. Tech programme in Bioengineering department at National Institute of Technology Agartala, India. Her research interests include molecular docking and simulations. PD is presently working as an Assistant Professor at Civil Engineering department at The ICFAI University, Tripura, India. His research interests include Pattern recognition, machine learning, and artificial intelligence. DD is presently working as an Assistant Professor at Bioengineering department at National Institute of Technology Agartala, India. Her research interests include proteomics and bioinformatics.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
The work described has not been published before and contains original computational data. The manuscript is not under consideration for publication anywhere else. The publication has been approved by all co-authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Garai, S., Thomas, J., Dey, P. et al. LGBM-ACp: an ensemble model for anticancer peptide prediction and in silico screening with potential drug targets. Mol Divers (2023). https://doi.org/10.1007/s11030-023-10602-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-023-10602-0