Abstract
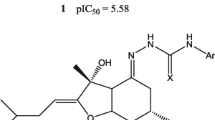
We have performed the docking of sulfonyl hydrazides complexed with cytosolic branched-chain amino acid aminotransferase (BCATc) to study the orientations and preferred active conformations of these inhibitors. The study was conducted on a selected set of 20 compounds with variation in structure and activity. In addition, the predicted inhibitor concentration (IC50) of the sulfonyl hydrazides as BCAT inhibitors were obtained by a quantitative structure–activity relationship (QSAR) method using three-dimensional (3D) vectors. We found that three-dimensional molecule representation of structures based on electron diffraction (3D-MoRSE) scheme contains the most relevant information related to the studied activity. The statistical parameters [cross-validate correlation coefficient (Q 2 = 0.796) and fitted correlation coefficient (R 2 = 0.899)] validated the quality of the 3D-MoRSE predictive model for 16 compounds. Additionally, this model adequately predicted four compounds that were not included in the training set.
Similar content being viewed by others
References
Ichihara A (1985) Aminotransferases of branched-chain amino acids. In: Christen P, Metzler DE (eds) Transaminases. Wiley, New York, pp 430–439
Bixel MG, Hutson SM, Hamprecht B (1997) Cellular distribution of branched-chain amino acid aminotransferase isoenzymes among rat brain glial cells in culture. J Histochem Cytochem 45: 685–694
Hutson SM, Fenstermacher D, Mahar C (1988) Role of mitochondrial transamination in branched chain amino acid metabolism. J Biol Chem 263: 3618–3625
Hutson SM, Wallin R, Hall TR (1992) Identification of mitochondrial branched chain aminotransferase and its isoforms in rat tissues. J Biol Chem 267: 15681–15686
Hall TR, Wallin R, Reinhart GD, Hutson SM (1993) Branched chain aminotransferase isoenzymes. J Biol Chem 268: 3092–3098
Lieth E, LaNoue K, Berkich D, Xu B, Ratz M, Taylor C, Hutson SM (2001) Nitrogen shuttling between neurons and glial cells during glutamate synthesis. J Neurochem 76: 1712–1723. doi:10.1046/j.1471-4159.2001.00156.x
Hutson S (2001) Structure and function of branched chain aminotransferases. Prog Nucleic Acid Res Mol Biol 70: 175–206. doi:10.1016/S0079-6603(01)70017-7
Hays SJ, Hu L-Y, Lei H, Scholten JD, Wustrow DJ (2002) US Patent 6632831
Hutson SM, Berkich DA, Drown P, Xu B, LaNoue KF (1998) Role of branched-chain aminotransferase isoenzymes and gabapentin in neurotransmitter metabolism. J Neurochem 71: 863–874
Hu L-Y, Boxer PA, Kesten SR, Lei HJ, Wustrow DJ, Moreland DW, Zhang L, Ahn K, Ryder TR, Liu X, Rubin JR, Fahnoe K, Carroll R, Dutta T, Fahnoe S, Probert DC, Roof AW, Rafferty RL, Kostlan MF, Scholten CR, Hood JD, Ren M, Schielke X-D, Su GP, Taylor T-Z, Mistry CP, Mc A, Connell P, Hasemann C, Ohren J (2006) The design and synthesis of human branched-chain amino acid aminotransferase inhibitors for treatment of neurodegenerative diseases. Bioorg Med Chem Lett 16: 2337–2340. doi:10.1016/j.bmcl.2005.07.058
Sekhar PN, Amrutha RN, Sangam S, Verma DPS, Kishor PBK (2007) Biochemical characterization, homology modeling and docking studies of ornithine δ-aminotransferase—an important enzyme in proline biosynthesis of plants. J Mol Graph Model 26: 709–719. doi:10.1016/j.jmgm.2007.04.006
Seo J-H, Park H-Y, Kim J, Lee B-S, Kim B-G (2008) Exploring sequence space: profile analysis and protein–ligand docking to screen ω-aminotransferases with expanded substrate specificity. Biotechnol J 3: 676–686. doi:10.1002/biot.200700264
Pârvu L (2003) QSAR—a piece of drug design. J Cell Mol Med 7: 333–335. doi:10.1111/j.1582-4934.2003.tb00235.x
Dewar MJS, Thiel W (1977) Ground states of molecules. 38. The MNDO method. Approximations and parameters. J Am Chem Soc 99: 4899–4907. doi:10.1021/ja00457a004
Biosym Technologies (1993) Insight II Version 2.3, Discover Version 2.9.5 Biosym Technologies, San Diego, USA
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19: 1639–1662. doi:10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B
Tripos Inc. (2006) SYBYL version 7.3, Tripos Inc., 1699 South Hanley Road, St. Louis, MO 63144, USA
Todeschini R, Consonni V, Pavan M (2002) Dragon Software version 2.1
Hemmer MC, Steinhauer V, Gasteiger J (1999) Deriving the 3D structure of organic molecules from their infrared spectra. Vib Spectrosc 19: 151–164. doi:10.1016/S0924-2031(99)00014-4
Hemmer MC, Gasteiger J (2000) Prediction of three-dimensional molecular structures using information from infrared spectra. Anal Chim Acta 420: 145–154. doi:10.1016/S0003-2670(00)00876-X
Schuur J, Selzer P, Gasteiger J (1996) The coding of the three-dimensional structure of molecules by molecular transforms and its application to structure-spectra correlations and studies of biological activity. J Chem Inf Comput Sci 36: 334–344. doi:10.1021/ci950164c
Todeschini R, Lansagni M, Marengo E (1994) New molecular descriptors for 2D and 3D structures theory. J Chemometr 8: 263–272. doi:10.1002/cem.1180080405
Consonni V, Todeschini R, Pavan M (2002) Structure/response correlations and similarity/diversity analysis by GETAWAY descriptors: 1. Theory of the novel 3D molecular descriptors. J Chem Inf Comput Sci 42: 682–692. doi:10.1021/ci015504a
The Mathworks Inc. (2004) MATLAB version 7.0. The Mathworks Inc., Natick, MA, http://www.mathworks.com
Cronin MTD, Schultz TW (2003) Pitfalls in QSAR. J Mol Struct THEOCHEM 622: 39–51. doi:10.1016/S0166-1280(02)00616-4
Fernández M, Tundidor-Camba A, Caballero J (2005) Modeling of cyclin-dependent kinase inhibition by 1H-pyrazolo [3,4-d] pyrimidine derivatives using artificial neural network ensembles. J Chem Inf Model 45: 1884–1895. doi:10.1021/ci050263i
Caballero J, Fernández M (2006) Linear and nonlinear modeling of antifungal activity of some heterocyclic ring derivatives using multiple linear regression and Bayesian-regularized neural networks. J Mol Model 12: 168–181. doi:10.1007/s00894-005-0014-x
Fernández M, Caballero J (2006) Ensembles of Bayesian-regularized genetic neural networks for modeling of acetylcholinesterase inhibition by huprines. Chem Biol Drug Des 68: 201–212. doi:10.1111/j.1747-0285.2006.00435.x
Fernández M, Carreiras MC, Marco JL, Caballero J (2006) Modeling of acetylcholinesterase inhibition by tacrine analogues using Bayesian–regularized Genetic Neural Networks and ensemble averaging. J Enzyme Inhib Med Chem 21: 647–661. doi:10.1080/14756360600862366
Duchowicz PR, Fernández M, Caballero J, Castro EA, Fernández FM (2006) QSAR for non-nucleoside inhibitors of HIV-1 reverse transcriptase. Bioorg Med Chem 14: 5876–5889. doi:10.1016/j.bmc.2006.05.027
Caballero J, Fernández M (2008) Artificial neural networks from MATLAB® in medicinal chemistry. Bayesian-regularized genetic neural networks (BRGNN): Application to the prediction of the antagonistic activity against human platelet thrombin receptor (PAR-1). Curr Top Med Chem 8: 1580–1605. doi:10.2174/156802608786786570
Saíz-Urra L, González MP, Teijeira M (2006) QSAR studies about cytotoxicity of benzophenazines with dual inhibition toward both topoisomerases I and II: 3D-MoRSE descriptors and statistical considerations about variable selection. Bioorg Med Chem 14: 7347–7358. doi:10.1016/j.bmc.2006.05.081
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Caballero, J., Vergara-Jaque, A., Fernández, M. et al. Docking and quantitative structure–activity relationship studies for sulfonyl hydrazides as inhibitors of cytosolic human branched-chain amino acid aminotransferase. Mol Divers 13, 493–500 (2009). https://doi.org/10.1007/s11030-009-9140-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-009-9140-1