Abstract
Insect embryonic development and morphology are characterized by their anterior–posterior and dorsal–ventral (DV) patterning. In Drosophila embryos, DV patterning is mediated by a dorsal protein gradient which activates twist and snail proteins, the important regulators of DV patterning. To activate or repress gene expression, some regulatory proteins bind in clusters to their target gene at sites known as cis-regulatory elements or enhancers. To understand how variations in gene expression in different lineages might lead to different phenotypes, it is necessary to understand enhancers and their evolution. Drosophila melanogaster has been widely studied to understand the interactions between transcription factors and the transcription factor binding sites. Tribolium castaneum is an upcoming model animal which is catching the interest of biologists and the research on the enhancer mechanisms in the insect’s axes patterning is still in infancy. Therefore, the current study was designed to compare the enhancers of DV patterning in the two insect species. The sequences of ten proteins involved in DV patterning of D. melanogaster were obtained from Flybase. The protein sequences of T. castaneum orthologous to those obtained from D. melanogaster were acquired from NCBI BLAST, and these were then converted to DNA sequences which were modified by adding 20 kb sequences both upstream and downstream to the gene. These modified sequences were used for further analysis. Bioinformatics tools (Cluster–Buster and MCAST) were used to search for clusters of binding sites (enhancers) in the modified DV genes. The results obtained showed that the transcription factors in Drosophila melanogaster and Tribolium castaneum are nearly identical; however, the number of binding sites varies between the two species, indicating transcription factor binding site evolution, as predicted by two different computational tools. It was observed that dorsal, twist, snail, zelda, and Supressor of Hairless are the transcription factors responsible for the regulation of DV patterning in the two insect species.












Similar content being viewed by others
Data availability
All the data has been made available.
References
Berni M, Fontenele MR, Tobias-Santos V, Caceres-Rodrigues A, Mury FB, Vionette-do-Amaral R, Masuda H, Sorgine M, da Fonseca RN, Araujo H (2014) Toll signals regulate dorsal–ventral patterning and anterior–posterior placement of the embryo in the hemipteran Rhodnius prolixus. EvoDevo 5(1):8. https://doi.org/10.1186/2041-9139-5-38
Nei M (2007) The new mutation theory of phenotypic evolution. PNAS 104:12235–12242. https://doi.org/10.1073/pnas.0703349104
Moses AM, Pollard DA, Nix DA, Iyer VN, Li X-Y, Biggin MD, Eisen MB (2006) Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol 2:130. https://doi.org/10.1371/journal.pcbi.0020130
Lettice LA, Heaney SJH, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE (2003) A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12:1725–1735. https://doi.org/10.1093/hmg/ddg180
Vijayabaskar MS, Goode DK, Obier N, Lichtinger M, Emmett AML, Abidin FNZ, Shar N, Hannah R, Assi SA, Lie-A-Ling M, Gottgens B, Lacaud G, Kouskoff V, Bonifer Constanze, Westhead DR (2019) Identification of gene specific cis-regulatory elements during differentiation of mouse embryonic stem cells: an integrative approach using high-throughput datasets. PLoS Comput Biol 15:e1007337. https://doi.org/10.1371/journal.pcbi.1007337
Carroll SB (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134:25–36. https://doi.org/10.1016/j.cell.2008.06.030
Spitz F, Furlong EE (2012) Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13:613–626. https://doi.org/10.1038/nrg3207
Lai YT, Deem KD, Borràs-Castells F, Sambrani N, Rudolf H, Suryamohan K, El-Sherif E, Halfon MS, McKay DJ, Tomoyasu Y (2018) Enhancer identification and activity evaluation in the red flour beetle, Tribolium castaneum. Development 145(7):dev160663
Paris M, Kaplan T, Li XY, Villalta JE, Lott SE, Eisen MB (2013) Extensive divergence of transcription factor binding in drosophila embryos with highly conserved gene expression. PLoS Genet 9:e1003748. https://doi.org/10.1371/journal.pgen.1003748
He Q, Bardet AF, Patton B, Purvis J, Johnston J et al (2011) High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet 43:414–420
Bolognesi R, Beermann A, Farzana L, Wittkopp N, Lutz R, Balavoine G, Brown SJ, Schroder R (2008) Tribolium Wnts: evidence for a larger repertoire in insects with overlapping expression patterns that suggest multiple redundant functions in embryogenesis. Dev Genes Evol 218:193–202. https://doi.org/10.1007/s00427-007-0170-3
Martin BL, Kimelman D (2009) Wnt signaling and the evolution of embryonic posterior development. Curr Biol 19:R215-219. https://doi.org/10.1016/j.cub.2009.01.052
Darras S, Fritzenwanker JH, Uhlinger KR, Farrelly E, Pani AM, Hurley IA, Norris RP, Osovitz M, Terasaki M, Wu M, Aronowicz J, Kirschner M, Gerhart JC, Lowe CJ (2018) Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol 16:e2003698. https://doi.org/10.1371/journal.pbio.2003698
Petersen CP, Reddien PW (2009) Wnt signaling and the polarity of the primary body axis. Cell 139:1056–1068. https://doi.org/10.1016/j.cell.2009.11.035
De Robertis EM (2008) Evo-Devo: variations on ancestral themes. Cell 132:185–195. https://doi.org/10.1016/j.cell.2008.01.003
Lynch JA, Roth S (2011) The evolution of dorsal-ventral patterning mechanisms in insects. Genes Dev 25:107–118. https://doi.org/10.1101/gad.2010711
Sachs L, Chen YT, Drechsler A, Lynch JA, Panfilio KA, Lassig M, Berg J, Roth S (2015) Dynamic BMP signaling polarized by Toll patterns the dorsoventral axis in a hemimetabolous insect. eLife 4:5502
O’Connor MB, Umulis D, Othmer HG, Blair SS (2006) Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development 133:183–193. https://doi.org/10.1242/dev.02214
Nunes da Fonseca R, van der Zee M, Roth S (2010) Evolution of extracellular Dpp modulators in insects: the roles of tolloid and twisted-gastrulation in dorsoventral patterning of the Tribolium embryo. Dev Biol 345:80–93. https://doi.org/10.1016/j.ydbio.2010.05.019
Bressan D, Araujo HM (2022) Evolution of the dorsoventral axis in insects: the changing role of Bone Morphogenetic Proteins. Curr Opin Insect Sci 49:1–7
Kapil S, Kaur T (2021) Computational prediction and analysis of Dorsal-ventral patterning gene enhancers in Drosophila melanogaster. UPJOZ 42:83–89
Small S, Arnosti DN (2020) Transcriptional Enhancers in Drosophila. Genetics 216:1–26. https://doi.org/10.1534/genetics.120.301370
Tribolium Genome Sequencing Consortium (2008) The genome of the model beetle and pest Tribolium castaneum. Nature 452:949–955. https://doi.org/10.1038/nature06784
Pechmann M, Kenny NJ, Pott L, Heger P, Chen YT, Buchta T, Ozuak O, Lynch JA, Roth S (2021) Striking parallels between dorsoventral patterning in Drosophila and Gryllus reveal a complex evolutionary history behind a model gene regulatory network. eLife 10:e68287. https://doi.org/10.7554/eLife.68287
Brown SJ, Denell RE (1996) Segmentation and dorsal–ventral patterning in Tribolium. Semin Cell Dev Biol 7:553–560. https://doi.org/10.1006/scdb.1996.0069
Handel K, Basal A, Fan X, Roth S (2005) Tribolium castaneum twist: gastrulation and mesoderm formation in a short-germ beetle. Dev Genes Evol 215:13–31. https://doi.org/10.1007/s00427-004-0446-9
Keller R (2002) Shaping the vertebrate body plan by polarized embryonic cell movements. Science 298:1950–1954. https://doi.org/10.1126/science.1079478
Kong D, Wolf F, Großhans J (2017) Forces directing germ-band extension in Drosophila embryos. Mech Dev 144:11–22. https://doi.org/10.1016/j.mod.2016.12.001
Gerttula S, Jin YS, Anderson KV (1988) Zygotic expression and activity of the Drosophila Toll gene, a gene required maternally for embryonic dorsal-ventral pattern formation. Genetics 119:123–133. https://doi.org/10.1093/genetics/119.1.123
Zee M, Stockhammer O, von Levetzow C, Nunes da Fonseca R, Roth S (2006) Sog/Chordin is required for ventral-to-dorsal Dpp/BMP transport and head formation in a short germ insect. PNAS 103:16307–16312. https://doi.org/10.1073/pnas.0605154103
El-Sherif E, Zhu X, Fu J, Brown SJ (2014) Caudal regulates the spatiotemporal dynamics of pair-rule waves in Tribolium. PLoS Genet 1:e1004677. https://doi.org/10.1371/journal.pgen.1004677
Cande J, Goltsev Y, Levine MS (2009) Conservation of enhancer location in divergent insects. Proc Natl Acad Sci USA 106:14414–14419. https://doi.org/10.1073/pnas.0905754106
Moussian B, Roth S (2005) Dorsoventral axis formation in the Drosophila embryo shaping and transducing a morphogen gradient. Curr Biol 15:R887–R899. https://doi.org/10.1016/j.cub.2005.10.026
Stappert D, Frey N, von Levetzow C, Roth S (2016) Genome-wide identification of Tribolium dorsoventral patterning genes. Development 143:2443–2454. https://doi.org/10.1242/dev.130641
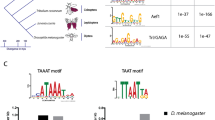
Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Cheneby J, Kulkarni SR, Tan G, Baranasic D, Arenillas DJ, Sandelin A, Vandepoele K, Lenhard B, Ballester B, Wasserman WW, Parcy F, Mathelier A (2018) JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46:D260–D266. https://doi.org/10.1093/nar/gkx1126
Ozdemir A, Ma L, White KP, Stathopoulos A (2014) Su (H)-mediated repression positions gene boundaries along the dorsal-ventral axis of Drosophila embryos. Dev Cell 31:100–113. https://doi.org/10.1016/j.devcel.2014.08.005
Zinzen RP, Senger K, Levine M, Papatsenko D (2006) Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol 16:1358–1365. https://doi.org/10.1016/j.cub.2006.05.044
Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294:1519–1521. https://doi.org/10.1126/science.1065201
Frith MC, Li MC, Weng Z (2003) Cluster-Buster: finding dense clusters of motifs in DNA sequences. Nucleic Acids Res 31:3666–3668. https://doi.org/10.1093/nar/gkg540
Bailey TL, Noble WS (2003) Searching for statistically significant regulatory modules. Bioinformatics 19:16–25. https://doi.org/10.1093/bioinformatics/btg1054
Morisato D, Anderson KV (1995) Signaling pathways that establish the dorsal-ventral pattern of the Drosophila embryo. Annu Rev Genet 29:371–399. https://doi.org/10.1146/annurev.ge.29.120195.002103
Jiang J, Kosman D, Ip YT, Levine M (1991) The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev 5:1881–1891
Goltsev Y, Fuse N, Frasch M, Zinzen RP, Lanzaro G, Levine M (2007) Evolution of the dorsal-ventral patterning network in the mosquito, Anopheles gambiae. Development 134:2415–2424. https://doi.org/10.1242/dev.02863
Stathopoulos A, Levine M (2004) Whole-genome analysis of Drosophila gastrulation. Curr Opin Genet Dev 14:477–484. https://doi.org/10.1016/j.gde.2004.07.004
Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M (1992) dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev 6:1518–1530
Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M (2002) Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell 111:687–701. https://doi.org/10.1016/S0092-8674(02)01087-5
Stathopoulos A, Levine M (2005) Genomic regulatory networks and animal development. Dev Cell 9:449–462. https://doi.org/10.1016/j.devcel.2005.09.005
Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M (2007) Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev 21:385–390. https://doi.org/10.1101/gad.1509607
Sandler JE, Stathopoulos A (2016) Stepwise progression of embryonic patterning. Trends Genet 32:432–443. https://doi.org/10.1016/j.tig.2016.04.004
Papadopoulos DK, Tomancak P (2019) Gene regulation: analog to digital conversion of transcription factor gradients. Curr Biol 29:R422–R424. https://doi.org/10.1016/j.cub.2019.04.052
Hong JW, David A, Hendrix LM (2008) Shadow enhancers as a source of evolutionary novelty. Science 321:1314. https://doi.org/10.1126/science.1160631
Garcia M, Stathopoulos A (2011) Lateral gene expression in Drosophila early embryos is supported by grainyhead-mediated activation and tiers of dorsally-localized repression. PLoS ONE 6:9172. https://doi.org/10.1371/journal.pone.0029172
Von Ohlen TL, Harvey C, Panda M (2007) Identification of an upstream regulatory element reveals a novel requirement for Ind activity in maintaining ind expression. Mech Dev 124:230–236. https://doi.org/10.1016/j.mod.2006.11.003
Wheeler SR, Carrico ML, Wilson BA, Skeath JB (2005) The Tribolium columnar genes reveal conservation and plasticity in neural precursor patterning along the embryonic dorsal–ventral axis. Dev Biol 279:491–500. https://doi.org/10.1016/j.ydbio.2004.12.031
Stauber M, Prell A, Schmidt-Ott U (2002) A single Hox3 gene with composite bicoid and zerknüllt expression characteristics in non-Cyclorrhaphan flies. PNAS 99:274–279. https://doi.org/10.1073/pnas.012292899
Rushlow C, Frasch M, Doyle H, Levine M (1987) Maternal regulation of zerknüllt: a homoeobox gene controlling differentiation of dorsal tissues in Drosophila. Nature 330:583–586. https://doi.org/10.1038/330583a0
Doyle HJ, Kraut R, Levine M (1989) Spatial regulation of zerknullt: A dorsal-ventral patterning gene in Drosophila. Genes Dev 3:1518–1533. https://doi.org/10.1101/gad.3.10.1518
Mellerick DM, Nirenberg M (1995) Dorsal-ventral patterning genes restrict NK-2 homeobox gene expression to the ventral half of the central nervous system of Drosophila embryos. Dev Biol 171:306–316. https://doi.org/10.1006/dbio.1995.1283
Jimenez F, Martin-Morris LE, Velasco L, Chu H, Sierra J, Rosen DR, White K (1995) vnd, a gene required for early neurogenesis of Drosophila, encodes a homeodomain protein. EMBO J 14:3487–3495. https://doi.org/10.1002/j.1460-2075.1995.tb07355.x
Erives A, Levine M (2004) Coordinate enhancers share common organizational features in the Drosophila genome. PNAS 101:3851–3856. https://doi.org/10.1073/pnas.0400611101
Morel V, Schweisguth F (2000) Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev 14:377–388. https://doi.org/10.1101/gad.14.3.377
Biemar F, David A, Nix PJ, Peterson B, Ronshaugen M, Sementchenko V, Bell I, Manak JR, Levine MS (2006) Comprehensive identification of Drosophila dorsal–ventral patterning genes using a whole-genome tiling array. PNAS 103:12763–12768. https://doi.org/10.1073/pnas.0604484103
Zhang JH, Levine M, Hilary A (2001) Brinker is a sequence-specific transcriptional repressor in the Drosophila embryo. Genes Dev 15:261–266. https://doi.org/10.1101/gad.861201
Markstein M, Markstein P, Markstein V, Levine MS (2002) Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci USA 99:763–768. https://doi.org/10.1073/pnas.012591199
Sommer RJ, Tautz D (1994) Expression patterns of twist and snail in Tribolium (Coleoptera) suggest a homologous formation of mesoderm in long and short germ band insects. Dev Genet 15:32–37. https://doi.org/10.1002/dvg.1020150105
Reeves GT, Stathopoulos A (2009) Graded dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb Perspect Biol 1:a000836
Sturtevant MA, Roark M, Bier E (1993) The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev 7:961–973. https://doi.org/10.1101/gad.7.6.96
Huang AM, Rusch J, Levine M (1997) An anteroposterior Dorsal gradient in the Drosophila embryo. Genes Dev 11:1963–1973. https://doi.org/10.1101/gad.11.15.1963
Bansal A, Upadhyay AK (2017) Genome-wide identification and analysis of putative Rhomboid Gene enhancers in multiple Drosophila species. Immunome Res 13:143. https://doi.org/10.4172/1745-7580.1000143
Acknowledgements
The authors would like to acknowledge the contributions of Dr. Rupinder Sayal, Ex. Assistant Professor, Department of Biochemistry, D.A.V. University, Jalandhar, in analyzing the results.
Funding
The authors have no relevant financial or non-financial interests to disclose.
Author information
Authors and Affiliations
Contributions
Subham Kapil has written the manuscript and made the figures, and Dr. Tejinder Kaur has supervised the work and edited the manuscript along with Dr. Ranbir Chander Sobti.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kapil, S., Sobti, R.C. & Kaur, T. Prediction and analysis of cis-regulatory elements in Dorsal and Ventral patterning genes of Tribolium castaneum and its comparison with Drosophila melanogaster. Mol Cell Biochem 479, 109–125 (2024). https://doi.org/10.1007/s11010-023-04712-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-023-04712-4