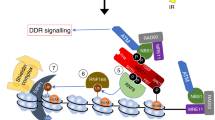
Abstract
DNA is continuously exposed to damaging agents that can lead to changes in the genetic information with adverse consequences. Nonetheless, eukaryotic cells have mechanisms such as the DNA damage response (DDR) to prevent genomic instability. The DNA of eukaryotic cells is packaged into nucleosomes, which fold the genome into highly condensed chromatin, but relatively little is known about the role of chromatin accessibility in DNA repair. p19INK4d, a cyclin-dependent kinase inhibitor, plays an important role in cell cycle regulation and cellular DDR. Extensive data indicate that p19INK4d is a critical factor in the maintenance of genomic integrity and cell survival. p19INK4d is upregulated by various genotoxics, improving the repair efficiency for a variety of DNA lesions. The evidence of p19INK4d translocation into the nucleus and its low sequence specificity in its interaction with DNA prompted us to hypothesize that p19INK4d plays a role at an early stage of cellular DDR. In the present study, we demonstrate that upon oxidative DNA damage, p19INK4d strongly binds to and relaxes chromatin. Furthermore, in vitro accessibility assays show that DNA is more accessible to a restriction enzyme when a chromatinized plasmid is incubated in the presence of a protein extract with high levels of p19INK4d. Nuclear protein extracts from cells overexpressing p19INK4d are better able to repair a chromatinized and damaged plasmid. These observations support the notion that p19INK4d would act as a chromatin accessibility factor that allows the access of the repair machinery to the DNA damage site.





Similar content being viewed by others
Abbreviations
- INK4:
-
Inhibitor of cyclin-dependent kinase 4
- CDK:
-
Cyclin-dependent kinase
- MNase:
-
Micrococcal nuclease
- DDR:
-
DNA damage response
- OGG1:
-
8-Oxoguanine DNA glycosylase 1
References
Levy-Wilson B, Fortier C, Blackhart BD, McCarthy BJ (1988) DNase I- and micrococcal nuclease-hypersensitive sites in the human apolipoprotein B gene are tissue specific. Mol Cell Biol 8:71–80
Friedberg EC (2003) DNA damage and repair. Nature 421:436–440
Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461:1071–1078
Barzilai A (2010) DNA damage, neuronal and glial cell death and neurodegeneration. Apoptosis 15:1371–1381
Meek DW (2009) Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer 9:714–723
Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411:366–374
Mallette FA, Gaumont-Leclerc MF, Ferbeyre G (2007) The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev 21:43–48
Zhivotovsky B, Kroemer G (2004) Apoptosis and genomic instability. Nat Rev Mol Cell Biol 5:752–762
Green CM, Almouzni G (2002) When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep 3:28–33
Polo SE, Almouzni G (2007) Chromatin dynamics during the repair of DNA lesions. Med Sci (Paris) 23:29–31
Soria G, Polo SE, Almouzni G (2012) Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell 46:722–734
Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, Almouzni G, Tora L (2001) UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J 20:3187–3196
Kikuchi M, Okumura F, Tsukiyama T, Watanabe M, Miyajima N, Tanaka J, Imamura M, Hatakeyama S (2009) TRIM24 mediates ligand-dependent activation of androgen receptor and is repressed by a bromodomain-containing protein, BRD7, in prostate cancer cells. Biochim Biophys Acta 1793:1828–1836
Loizou JI, Murr R, Finkbeiner MG, Sawan C, Wang ZQ, Herceg Z (2006) Epigenetic information in chromatin: the code of entry for DNA repair. Cell Cycle 5:696–701
Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z (2006) Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol 8:91–99
Rubbi CP, Milner J (2003) p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. EMBO J 22:975–986
Kuo WH, Wang Y, Wong RP, Campos EI, Li G (2007) The ING1b tumor suppressor facilitates nucleotide excision repair by promoting chromatin accessibility to XPA. Exp Cell Res 313:1628–1638
Roussel MF (1999) The INK4 family of cell cycle inhibitors in cancer. Oncogene 18:5311–5317
Pei XH, Xiong Y (2005) Biochemical and cellular mechanisms of mammalian CDK inhibitors: a few unresolved issues. Oncogene 24:2787–2795
Canepa ET, Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Sirkin PF, Ogara MF (2007) INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life 59:419–426
Al-Mohanna MA, Al-Khalaf HH, Al-Yousef N, Aboussekhra A (2007) The p16INK4a tumor suppressor controls p21WAF1 induction in response to ultraviolet light. Nucleic Acids Res 35:223–233
Ceruti JM, Scassa ME, Flo JM, Varone CL, Canepa ET (2005) Induction of p19INK4d in response to ultraviolet light improves DNA repair and confers resistance to apoptosis in neuroblastoma cells. Oncogene 24:4065–4080
Ceruti JM, Scassa ME, Marazita MC, Carcagno AC, Sirkin PF, Canepa ET (2009) Transcriptional upregulation of p19INK4d upon diverse genotoxic stress is critical for optimal DNA damage response. Int J Biochem Cell Biol 41:1344–1353
Varone CL, Canepa ET (1997) Evidence that protein kinase C is involved in delta-aminolevulinate synthase expression in rat hepatocytes. Arch Biochem Biophys 341:259–266
Mendez J, Stillman B (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 20:8602–8612
Frenster JH, Allfrey VG, Mirsky AE (1963) Repressed and active chromatin isolated from interphase lymphocytes. Proc Natl Acad Sci USA 50:1026–1032
Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T (2006) Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 10:105–116
Amouroux R, Campalans A, Epe B, Radicella JP (2010) Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res 38:2878–2890
Kamakaka RT, Bulger M, Kaufman PD, Stillman B, Kadonaga JT (1996) Postreplicative chromatin assembly by Drosophila and human chromatin assembly factor 1. Mol Cell Biol 16:810–817
Shaffer CD, Wuller JM, Elgin SC (1994) Raising large quantities of Drosophila for biochemical experiments. Methods Cell Biol 44:99–108
Bonte E, Becker PB (2009) Preparation of chromatin assembly extracts from preblastoderm Drosophila embryos. Methods Mol Biol 523:1–10
Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11:1475–1489
Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80:859–868
Ballmaier D, Epe B (2006) DNA damage by bromate: mechanism and consequences. Toxicology 221:166–171
Lindahl T, Karran P, Wood RD (1997) DNA excision repair pathways. Curr Opin Genet Dev 7:158–169
Barnes DE, Lindahl T (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 38:445–476
David SS, O’Shea VL, Kundu S (2007) Base-excision repair of oxidative DNA damage. Nature 447:941–950
Marazita MC, Ogara MF, Sonzogni SV, Marti M, Dusetti NJ, Pignataro OP, Canepa ET (2012) CDK2 and PKA mediated-sequential phosphorylation is critical for p19INK4d function in the DNA damage response. PLoS One 7:e35638
Ogara MF, Sirkin PF, Carcagno AL, Marazita MC, Sonzogni SV, Ceruti JM, Canepa ET (2013) Chromatin relaxation-mediated induction of p19INK4d increases the ability of cells to repair damaged DNA. PLoS One 8:e61143
Wang QE, Han C, Zhang B, Sabapathy K, Wani AA (2012) Nucleotide excision repair factor XPC enhances DNA damage-induced apoptosis by downregulating the antiapoptotic short isoform of caspase-2. Cancer Res 72:666–675
Povirk LF (1996) DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat Res 355:71–89
Dedon PC, Goldberg IH (1992) Free-radical mechanisms involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin, and calicheamicin. Chem Res Toxicol 5:311–332
Legarza K, Yang LX (2006) New molecular mechanisms of action of camptothecin-type drugs. Anticancer Res 26:3301–3305
Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, Zhang H, Kohn KW (2003) Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat Res 532:173–203
Dingwall C, Laskey RA (1991) Nuclear targeting sequences—a consensus? Trends Biochem Sci 16:478–481
Mattaj IW, Englmeier L (1998) Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem 67:265–306
Lim MA, Kikani CK, Wick MJ, Dong LQ (2003) Nuclear translocation of 3′-phosphoinositide-dependent protein kinase 1 (PDK-1): a potential regulatory mechanism for PDK-1 function. Proc Natl Acad Sci USA 100:14006–14011
Dinant C, Houtsmuller AB, Vermeulen W (2008) Chromatin structure and DNA damage repair. Epigenetics Chromatin 1:9
Papamichos-Chronakis M, Peterson CL (2013) Chromatin and the genome integrity network. Nat Rev Genet 14:62–75
Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, D’Urso A, Naar AM, Kingston R, Rippe K, Mostoslavsky R (2013) SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell 51:454–468
Lai W, Li H, Liu S, Tao Y (2013) Connecting chromatin modifying factors to DNA damage response. Int J Mol Sci 14:2355–2369
van Attikum H, Gasser SM (2005) ATP-dependent chromatin remodeling and DNA double-strand break repair. Cell Cycle 4:1011–1014
Peterson CL, Cote J (2004) Cellular machineries for chromosomal DNA repair. Genes Dev 18:602–616
Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X (2004) INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767–775
van Attikum H, Fritsch O, Hohn B, Gasser SM (2004) Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777–788
Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H (2010) The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol 190:741–749
Van Den Broeck A, Nissou D, Brambilla E, Eymin B, Gazzeri S (2012) Activation of a Tip60/E2F1/ERCC1 network in human lung adenocarcinoma cells exposed to cisplatin. Carcinogenesis 33:320–325
Sun Y, Jiang X, Chen S, Fernandes N, Price BD (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA 102:13182–13187
Harte MT, O’Brien GJ, Ryan NM, Gorski JJ, Savage KI, Crawford NT, Mullan PB, Harkin DP (2010) BRD7, a subunit of SWI/SNF complexes, binds directly to BRCA1 and regulates BRCA1-dependent transcription. Cancer Res 70:2538–2547
Drost J, Mantovani F, Tocco F, Elkon R, Comel A, Holstege H, Kerkhoven R, Jonkers J, Voorhoeve PM, Agami R, Del Sal G (2010) BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol 12:380–389
Acknowledgments
We thank Anna Campalans and Thierry Kortulewski for their help with confocal microscopy images. This work was supported by research grants from Agencia Nacional de Promoción Científica y Tecnológica (ANCYPT PICT0196), Universidad de Buenos Aires (UBACYT 284), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET PIP 567), Association de Recherche contre le Cancer (ARC n° PJA 20131200165 to JPR) and Ministerio de Ciencia, Tecnología e Innovación Productiva (MINCyT-ECOS A09B01), Argentina. Exchanges between the Argentinian and French laboratories were supported by French Ministry of Foreign Affairs and Argentine Ministry of Science and through an ECOS-Sud Project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sonzogni, S.V., Ogara, M.F., Castillo, D.S. et al. Nuclear translocation of p19INK4d in response to oxidative DNA damage promotes chromatin relaxation. Mol Cell Biochem 398, 63–72 (2015). https://doi.org/10.1007/s11010-014-2205-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-014-2205-1