Abstract
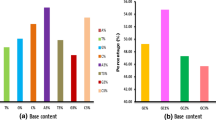
Comparison of ORFs between H. pylori strains 26695 and J99 showed that transitions (more than 3%) prevail over transversions (less than 1%). The predominance of transitions was explained by the high rates of cytosine replacement by thymine in the coding (3.5–5.3%) and noncoding (2.9–3.9%) DNA strands. The proportion of transversion-type correspondences (A → C, A → T, C → A, C → G, G → C, G → T, T → A, and T → G) did not exceed 0.84%. The highest proportion (28.3%) was observed for correspondences between C and T in ACGT-ATGT, the target site of active methyltransferase of H. pylori J99 (M.Hpy99XI). It was assumed that C → T mutations due to cytosine methylation-deamination are prevalent in H. pylori.
Similar content being viewed by others
REFERENCES
Marais A., Mendz G.L., Hazell S.L., Megraud F. 1999. Metabolism and genetics of Helicobacter pylori: The genome era. Microb. Mol. Biol. Rev. 63, 642–674.
Alm R.A., Ling L.S., Moir D.T., King B.L. et al. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 397, 176–180.
Tomb J.F., White O., Kerlavage A.R., Clayton R.A., et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 388, 539–547.
Alm R.A., Trust T.J. 1999. Analysis of the genetic diversity of Helicobacter pylori: The tale of two genomes. J. Mol. Med. 77, 834–846.
Boucher Y., Nesbo C.L., Doolittle W.F. 2001. Microbial genomes: Dealing with diversity. Curr. Opin. Microbiol. 4, 285–289.
Kuipers E., Israel D.A., Kusters J.G., Gerrits M.M., et al. 2000. Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host. J. Infect. Dis. 181, 273–282.
Israel D.A. Salama N., Krishna U., Rieger U.M., et al. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA. 98, 14625–14630.
Momynaliev K.T., Smirnova O.V., Kudryavtseva L.V., Govorun V.M. 2003. Comparative genome analysis of Helicobacter pylori strains. Mol. Biol. 37, 625–633.
Salama N., Guillemin K., McDaniel T.K., Sherlock G., Tompkins L., Falkow S. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA. 97, 14668–14673.
Kansau I., Raymond J., Bingen E., Courcoux P., et al. 1996. Genotyping of Helicobacter pylori isolates by sequencing of PCR products and comparison with the RAPD technique. Res. Microbiol. 147, 661–669.
Hook-Nikanne J., Berg D.E., Peek R.M. Jr., Kersulyte D., Tummuru M.K., Blaser M.J. 1998. DNA sequence conservation and diversity in transposable element IS605 of Helicobacter pylori. Helicobacter. 3, 79–85.
Suerbaum S, Smith J.M., Bapumia K., Morelli G., Smith N.H., et al. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA. 95, 12619–12624.
Garner J.A., Cover T.L. 1995. Analysis of genetic diversity in cytotoxin-producing and non-cytotoxin-producing Helicobacter pylori strains. J. Infect. Dis. 172, 290–293.
Evans Jr. D.J, Queiroz D.M., Mendes E.N., Evans D.G. 1998. Diversity in the variable region of Helicobacter pylori cagA gene involves more than simple repetition of a 102-nucleotide sequence. Biochem. Biophys. Res. Commun. 245, 780–784.
Aras R.A., Kang J., Tschumi A.I., Harasaki Y., Blaser M.J. 2003. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc. Natl. Acad. Sci. USA. 100, 13579–13584.
Wang G., Humayun M.Z., Taylor D.E. 1999. Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol. 7, 488–493.
Frederico L.A., Kunkel T.A., Shaw B.R. 1990. A sensitive genetic assay for the detection of cytosine deamination: Determination of rate constants and the activation energy. Biochemistry. 29, 2532–2537.
Shen J.-C., Rideout WM., Jones P.A. 1994. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 22, 972–976.
Coulondre C., Miller J.H., Farabaugh P.J., Gilbert W. 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 274, 775–780.
De Jong P.J., Grosvosky A.J. & Glickman B.W. 1988. Spectrum of spontaneous mutation at the APRT locus of Chinese hamster ovary cells: An analysis at the DNA sequence level. Proc. Natl. Acad. Sci. USA. 85, 3499–3503.
Halliday J.A., Glickman B.W. 1991. Mechanisms of spontaneous mutation in DNA repair-proficient Escherichia coli. Mutat. Res. 250, 55–71.
Douglas G.R., Gingerich J.D., Gossen J.A., Bartlett S.A. 1994. Sequence spectra of spontaneous lacZ gene mutations in transgenic mouse somatic and germline tissues. Mutagenesis. 9, 451–458.
Lindahl T., Nyberg B. 1974. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 13, 3405–3410.
Gromova E.S., Khoroshaev A.V. 2003. Prokaryotic DNA methyltransferases: Structure and mechanism of interaction with DNA. Mol. Biol. 37, 300–314.
Bell D.C., Cupples C.G. 2001. Very-short-patch repair in Escherichia coli requires the dam adenine methylase. J. Bacteriol. 183, 3631–3635.
Bhagwat A.S., Lieb M. 2002. Cooperation and competition in mismatch repair: Very short-patch repair and methyl-directed mismatch repair in Escherichia coli. Mol. Microbiol. 44, 1421–1428.
Suerbaum S., Smith J.M., Bapumia K., et al. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA. 95, 12619–12624.
Suerbaum S., Achtman M. 1999. Evolution of Helicobacter pylori: The role of recombination. Trends Microbiol. 7, 182–184.
Suerbaum S. 2000. Genetic variability within Helicobacter pylori. Int. J. Med. Microbiol. 290, 175–181.
Achtman M., Suerbaum S. 2000. Sequence variation in Helicobacter pylori. Trends Microbiol. 8, 57–58.
Suerbaum S., Achtman M. 2004. Helicobacter pylori: Recombination, population structure and human migrations. Int. J. Med. Microbiol. 294, 133–139.
Francino M.P., Ochman H. 1997. Strand asymmetries in DNA evolution. Trends Genet. 13, 240–245.
Cebrat S., Dudek M.R., Mackiewicz P., et al. 1997. Asymmetry of coding versus noncoding strand in coding sequences of different genomes. Microb. Comp. Genomics. 2, 259–268.
Rogerson A.C. 1989. The sequence asymmetry of the Escherichia coli chromosome appears to be independent of strand or function and may be evolutionarily conserved. Nucleic Acids Res. 17, 5547–5563.
Kowalczuk M., Mackiewicz P., Mackiewicz D., et al. 2001. DNA asymmetry and the replicational mutational pressure. J. Appl. Genet. 42, 553–577.
Nikolaou C., Almirantis Y. 2003. Mutually symmetric and complementary triplets: Differences in their use distinguish systematically between coding and non-coding genomic sequences. J. Theor. Biol. 223, 477–487.
Karlin S., Doerfler W., Cardon L.R. 1994. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J. Virol. 68, 2889–2897.
Karlin S., Mrazek J., Campbell A.M. 1997. Compositional biases of bacterial genomes and evolutionary implications. J. Bacteriol. 179, 3899–3913.
Breslauer K.J., Frank E., Blocker H., Marky L.A. 1986. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA. 83, 3746–3750.
Delcourt S.G., Blake R.D. 1991. Stacking energies in DNA. J. Biol. Chem. 266, 15160–15169.
Karlin S. Global dinucleotide signatures and analysis of genomic heterogeneity. 1998. Curr Opin. Microbiol. 1, 598–610.
Xia X. DNA methylation and Mycoplasma genomes. 2003. J. Mol. Evol. 57, 21–28.
Lin L.F., Posfai J., Roberts R.J., Kong H. 2001. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl. Acad. Sci. USA. 98, 2740–2745.
Kusano K., Naito T., Handa N., Kobayashi I. 1995. Restriction-modification systems as genomic parasites in competition for specific sequences. Proc. Natl. Acad. Sci. USA. 92, 11095–11099.
Author information
Authors and Affiliations
Additional information
__________
Translated from Molekulyarnaya Biologiya, Vol. 39, No. 6, 2005, pp. 945–951.
Original Russian Text Copyright © 2005 by Momynaliev, Rogov, Govorun.
Rights and permissions
About this article
Cite this article
Momynaliev, K.T., Rogov, S.I. & Govorun, V.M. Nucleotide Correspondence between Protein-Coding Sequences of Helicobacter pylori Strains 26695 and J99. Mol Biol 39, 826–832 (2005). https://doi.org/10.1007/s11008-005-0101-1
Received:
Issue Date:
DOI: https://doi.org/10.1007/s11008-005-0101-1