Abstract
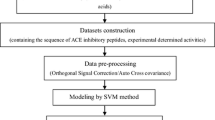
A quantitative multidimensional amino acids descriptors E (E1–E5) has been introduced in bioactive peptides Quantitative Structure–Activity Relationship (QSAR) study. These descriptors correlate well with hydrophobicity, size, preferences for amino acids to occur in α-helices, composition and the net charge, respectively. They were then applied to construct characterization and QSAR analysis on 48 angiotensin-converting enzyme (ACE) inhibitors dipeptides, 55 ACE inhibitors tripeptides and 48 bitter tasting dipeptides by support vector regression (SVR). The leave one out cross validation Q 2(CV) were 0.886, 0.985 and 0.912, the root mean square error (RMSE) were 0.250, 0.021 and 0.123, respectively. The results showed that, in comparison with the conventional descriptors, the new descriptor (E) is a useful structure characterization method for peptide QSAR analysis. The importance of each parameter or property at each position in peptides is estimated by the value of the model RMSE obtained using leave-one-parameter-out (LOPO) approach in the SVR model. This will be provided with certain guidance meaning to design and exploit peptide analogues. The results also indicate that SVR can be used as an alternative powerful modeling tool for peptide QSAR studies, and give one advice (LOPO) about evaluating the importance of parameter in SVR model. Moreover, it also offered an idea about nonlinear relation between bioactive of peptides and their structural descriptors E. The establishment of such methods will be a very meaningful work to peptide bioactive investigation in peptide analogue drug design.






Similar content being viewed by others
Abbreviations
- QSAR:
-
Quantitative structure–activity relationship
- ACE:
-
Angiotensin-converting enzyme
- LOPO:
-
Leave-one-parameter-out
- SVR:
-
Support vector regression
- MLR:
-
Multiple linear regression
- PLS:
-
Partial least square
- PCR:
-
Princupal component regression
- RMSE:
-
Root mean square error
- BA:
-
Biological activities
- LOOCV:
-
Leave-one-out cross-validation
References
Alex JS, Bernhrd S (2004) A tutorial on support vector regression. Stat Comput 14:199–222
Asao M, Iwamara H, Akamatsu M, Fujita T (1987) Quantitative structure–activity relationships of bitter thresholds of amino acids, peptides and their derivatives. J Med Chem 30(10):1873–1879
Cocchi M, Johansson E (1993) Amino acids characterization by GRID and multivariate data analysis. Quant Struct-Act Relat 12:1–8
Collantes ER, Dunn WJ (1995) Amino acid side chain descriptors for quantitative structure-activity relationship studies of peptide analogues. J Med Chem 38:2705–2713
Cushman DW, Cheung HS, Sabo EF, Ondetti MA (1981) Angiotensin converting enzyme inhibitors: evolution of a new class of antihypertensive drugs. In: Horovitz ZP (ed) Angiotensin converting enzyme inhibitors, mechanisms of action and clinical implications. Urban and Schwarzenberg, Baltimore, pp 3–25
Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikström C, Wold S (2001) Multi- and megavariate data analysis: principle and application. Umetrics AB, Umea, Sweden
Fan RE, Chen PH, Lin CJ (2005) Working set selection using the second order information for training SVM. J Mach Learn Res 6:1889–1918 or http://www.csie.ntu.edu.tw/~cjlin/libsvm/
Fang Y, Guo Y, Feng Y, Li M (2008) Predicting DNA-binding proteins: approached from Chou’s pseudo amino acid composition and other specific sequence features. Amino Acids 34(1):103–109
Guo Y, Yu L, Wen Z, Li M (2008) Using support vector machine combined with auto covariance to predict protein-protein interactions from protein sequences. Nucleic Acids Res 36(9):3025–3030
Hellberg S, Sjostrom M, Wold S (1986) The prediction of bradykinin potentiating potency of pentapeptides: an example of a peptide quantitative structure-activity relationship. Acta Chem Scand 40:135–140
Hellberg S, Sjostrom M, Skagerberg B, Wold S (1987) Peptide quantitative structure-activity relationships, a multivariate approach. J Med Chem 30:1126–1135
Hellberg S, Eriksson L, Jonsson J, Lindgren F, Sjostrom M, Skagerberg B, Wold S, Andrews P (1991) Minimum analogue peptide sets (MAPS) for quantitative structure-activity relationships. Int J Pept Protein Res 37:414–424
Jonsson J, Eriksson L, Hellberg S, Sjostrom M, Wold S (1989) Multivariate parameterization of 55 coded and non-coded amino acids. Quant Struct-Act Relat 8:204–209
Kidera A, Konishi Y, Oka M, Ooi T, Scheraga HA (1985) A statistical analysis of the physical properties of the 20 naturally occurring amino acids. J Protein Chem 4:23–55
Li SZ, Fu BH, Wang YQ, Liu SS (2001) On structural parameterization and molecular modeling of peptide analogues by molecular electronegativity-edge vector (VMEE): estimation and prediction for biological activity of pentapeptides. J Chin Chem Soc 48:937–944
Liang GZ, Zhou P, Zhou Y, Zhang QX, Li ZL (2006) New descriptors of amino acids and their applications to peptide quantitative structure activity relationship. Acta Chim Sinica 64(5):393–396
Lin ZH, Long HX, Bo Z, Wang YQ, Wu YZ (2008) New descriptors of amino acids and their application to peptide QSAR study. Peptides 28:1798–1805
Liu SS, Yin CS, Cai SX, Li ZL (2001) A novel MHDV descriptor for dipeptide QSAR studies. J Chin Chem Soc 48:253–260
Liu SS, Yin CS, Wang LS (2002) Combined MEDV-GA-MLR method for QSAR of three panels of steroids, dipeptides, and COX-2 indihibitors. J Chem Inf Comput Sci 42:749–756
Mei H, Zhou Y, Sun LL, Li ZL (2004) A new descriptors of amino acid and its application in peptide QSAR. Acta Phys Chim Sin 20:821–825
Mei H, Liao ZH, Zhou Y, Li SSZ (2005) A new set of amino acid descriptors and its application in peptide QSARs. Biopolymers (peptide science) 80(6):775–786
Niu B, Lu WC, Yang SS, Cai YD, Li GZ (2007) Support vector machine for SAR/QSAR of phenethyl-amines. Acta Pharmacol Sin 28:1075–1086
Ramos De Armas R, Gonzalez-Diaz H, Molina R, Perez-Gonzalez M, Uriarte E (2004) Stochastic-based descriptors studying peptides biological properties: modeling the bitter tasting threshold of dipeptides. Bioorg Med Chem 12:4815–4822
Raychaudhury C, Banerjee A, Bag P, Roy S (1999) Topological shape and size of peptides: identification of potential allele specific helper T cell antigenic sites. J Chem Inf Comput Sci 39:248–254
Sandberg M, Eriksson L, Jonsson J, Sjostrom M, Wold S (1998) New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. J Med Chem 41:2481–2491
Sewald N, Jakubke HD (2002) Peptides: chemistry and biology. Wiley-Vch Verlag GmbH & Co. KGaA, Weinheim
Sneath PH (1966) Relations between chemical structure and biological activity in peptides. J Theor Biol 12:157–195
Tian FF, Zhou P, Li ZL (2007) T-Scale as a novel vector of topological descriptors for amino acids and its application in QSARs of peptides. J Mol Struct (Theochem) 830:106–115
Tong J, Liu S, Zhou P, Wu B, Li Z (2008) A novel descriptor of amino acids and its application in peptide QSAR. J Theor Biol 253:90–97
Vapnik VN (2004) Statistical learning theory. Publishing House of Electronics Industry, Beijing
Venkatarajan MS, Braun W (2001) New quantitative descriptors of amino acids based on multidimensional scaling of a large number of physical–chemical properties. J Mol Model 7:445–453
Wold S, Eriksson L, Hellberg S, Jonsson J, Sjostrom M, Skagerberg B, Wikstrom C (1987) Principal property values for six non-natural amino acids and their application to a structure-activity relationship for oxytocin peptide analogues. Can J Chem 65:1814–1820
Wu J, Aluko RE, Nakai S (2006) Structural requirements of angiotensin I-converting enzyme inhibitory peptides: quantitative structure–and–activity relationship study of di- and tri-peptides. J Agric Food Chem 54:732–738
Yin JJ, Diao YB, Wen ZN, Wang ZM, Li ML (2009) Study of peptides quantitative structure-activity relationship base on E descriptors by multiple linear regression. Interdiscip Sci Comput Life Sci (submitted)
Zaliani A, Gancia E (1999) MS-WHIM scores for amino acids: a new 3D description for peptide QSAR and QSPR studies. J Chem Inf Comput Sci 39:525–533
Zhou P, Zhou Y, Wu SR, Li B, Tian FF, Li ZL (2006) A new descriptor of amino acids based on the three-dimensional vector of atomic interaction field. Chin Sci Bull 51(5):524–529
Acknowledgment
This work was supported by the National Natural Science Foundation of China (No. 20775052) and by the “211 Engineering Double Support Plan” Foundation of Sichuan Agricultural University (Yaan, China).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yin, J., Diao, Y., Wen, Z. et al. Studying Peptides Biological Activities Based on Multidimensional Descriptors (E) Using Support Vector Regression. Int J Pept Res Ther 16, 111–121 (2010). https://doi.org/10.1007/s10989-010-9210-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-010-9210-3