Abstract
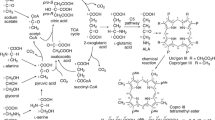
The biosynthetic pathways leading to δ-aminolevulinic acid (ALA) from the Shemin precursor glycine via the C5 pathway in Arthrobacter hyalinus were quantitatively evaluated by means of feeding experiments with [2-13C]glycine, sodium [1-13C]acetate, and sodium [2-13C]acetate, followed by analysis of the labeling patterns of coproporphyrinogen III (Copro’gen III) (biosynthesized from ALA) using 13C NMR spectroscopy. Two biosynthetic pathways leading to ALA from glycine via the C5 pathway were identified: i.e., transformation of glycine to l-serine catalyzed by glycine hydroxymethyltransferase, and glycine synthase-catalyzed catabolism of glycine to N 5,N 10-methylene-tetrahydrofolic acid (THF), which reacts with another molecule of glycine to afford l-serine. l-Serine is transformed to acetyl-CoA via pyruvic acid. Acetyl-CoA enters the tricarboxylic acid cycle, affording 2-oxoglutaric acid, which in turn is transformed to l-glutamic acid. The l-glutamic acid enters the C5 pathway, affording ALA in A. hyalinus. A 13C NMR spectroscopic comparison of the labeling patterns of Copro’gen III obtained after feeding of [2-13C]glycine, sodium [1-13C]acetate, and sodium [2-13C]acetate showed that [2-13C]glycine transformation and [2-13C]glycine catabolism in A. hyalinus proceed in the ratio of 52 and 48 %. The reaction of [2-13C]glycine and N 5,N 10-methylene-THF, that of glycine and N 5,N 10-[methylene-13C]methylene-THF generated from the [2-13C]glycine catabolism, and that of [2-13C]glycine and N 5,N 10-[methylene-13C]methylene-THF transformed the fed [2-13C]glycine to [1-13C]acetyl-CoA, [2-13C]acetyl-CoA, and [1,2-13C2]acetyl-CoA in the ratios of 42, 37, and 21 %, respectively. These labeled acetyl-CoAs were then incorporated into ALA. Our results provide a quantitative picture of the pathways of biosynthetic transformation to ALA from glycine in A. hyalinus.



Similar content being viewed by others
References
Shemin D, Kikuchi G (1958) Enzymatic synthesis of δ-aminolevulinic acid. Ann N Y Acad Sci 75:122–128
Kikuchi G, Kumar A, Talmage P, Shemin D (1958) The enzymatic synthesis of δ-aminolevulinic acid. J Biol Chem 233:1214–1219
Gibson KD, Laver WG, Neuberger A (1958) Initial stages in the biosynthesis of porphyrins. 2. The formation of δ-aminolaevulic acid from glycine and succinyl-coenzyme A by particles from chicken erythrocytes. Biochem J 70:71–81
Beale SI, Castelfranco PA (1974) Biosynthesis of δ-aminolevulinic acid in higher plants. II. Formation of δ-aminolevulinic acid from labeled precursors in greening plant tissues. Plant Physiol 53:297–303
Kannangara CG, Gough SP (1977) Biosynthesis of δ-aminolevulinate and chlorophyll by isolated chloroplasts. Carlsberg Res Commun 42:441–457
Kannangara CG, Gough SP (1978) Biosynthesis of δ-aminolevulinate in greening barley leaves: glutamate 1-semialdehyde aminotransferase. Carlsberg Res Commun 43:185–194
O’Neill GP, Chen M-W, Söll D (1989) δ-Aminolevulinic acid biosynthesis in Escherichia coli and Bacillus subtilis involves formation of glutamyl-tRNA. FEMS Microbiol Lett 60:255–260
Wider De Xifra EA, Del C. Batlle AM, Tigier HA (1971) δ-Aminolevulinate synthetase in extracts of cultured soybean cells. Biochim Biophys Acta 235:511–517
Ramaswamy NK, Nair PM (1973) δ-Aminolevulinic acid synthetase from cold-stored potatoes. Biochim Biophys Acta 293:269–277
Della Rosa RJ, Altman KI, Salomon K (1953) The biosynthesis of chlorophyll as studied with labeled glycine and acetic acid. J Biol Chem 202:771–779
Roberts DWA, Perkins HJ (1962) Chlorophyll biosynthesis and turnover in wheat leaves. Biochim Biophys Acta 58:499–506
Roberts DWA, Perkins HJ (1966) The incorporation of the two carbons of acetate and glycine into the phorbide and phytol moieties of chlorophylls a and b. Biochim Biophys Acta 127:42–46
Wellburn FAM, Wellburn AR (1971) Chlorophyll synthesis by isolated intact etioplasts. Biochem Biophys Res Commun 45:747–750
Beale SI, Castelfranco PA (1973) 14C Incorporation from exogenous compounds into δ-aminolevulinic acid by greening cucumber cotyledons. Biochem Biophys Res Commun 52:143–149
Okazaki T, Kurumaya K, Sagae Y, Kajiwara M (1990) Studies on biosynthesis of corrinoids and porphyrinoids. IV. Biosynthesis of chlorophyll in Euglena gracilis. Chem Pharm Bull 38:3303–3307
Oh-hama T, Seto H, Otake N, Miyachi S (1982) 13C-NMR evidence for the pathway of chlorophyll biosynthesis in green algae. Biochem Biophys Res Commun 105:647–652
Porra RJ, Klein O, Wrigh PE (1983) The proof by 13C-NMR spectroscopy of the predominance of the C5 pathway over the Shemin in chlorophyll biosynthesis in higher plants and of the formation of the methyl ester group of chlorophyll from glycine. Eur J Biochem 130:509–516
Kojima I, Maruhashi K, Sato H, Fujiwara Y (1993) A highly active producer of coproporphyrin III and uroporphyrin III. J Ferment Bioeng 76:527–529
Kajiwara M, Mizutani M, Matsuda R, Hara K, Kojima I (1994) A new biosynthetic pathway of porphyrins from isopropanol. J Ferment Bioeng 77:626–629
Iida K, Aoki T, Uegaki R, Kajiwara M (1997) Biosynthesis of corrinoids and porphyrinoids. XI. Source of oxaloacetic acid for uroporphyrinogen III biosynthesis in Arthrobacter hyalinus. Chem Pharm Bull 45:397–398
Kojima I, Maruhashi K, Fujiwara Y, Saito T, Kajiwara M, Mizutani M (1993) Identification of porphyrins produced from isopropanol by Arthrobacter hyalinus. J Ferment Bioeng 75:353–358
Acknowledgments
The author would like to thank Prof. Kajiwara for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Iida, K. Quantitative evaluation of the biosynthetic pathways leading to δ-aminolevulinic acid from the Shemin precursor glycine via the C5 pathway in Arthrobacter hyalinus by analysis of 13C-labeled coproporphyrinogen III biosynthesized from [2-13C]glycine, [1-13C]acetate, and [2-13C]acetate using 13C NMR spectroscopy. J Radioanal Nucl Chem 295, 1819–1827 (2013). https://doi.org/10.1007/s10967-012-2104-4
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10967-012-2104-4