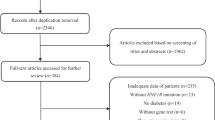
Abstract
Mutations in HNF transcription factor genes cause the most common subtypes of maturity-onset of diabetes of youth (MODY), a monogenic form of diabetes mellitus. Mutations in the HNF1-α, HNF4-α, and HNF1-β genes are primarily considered as the cause of MODY3, MODY1, and MODY5 subtypes, respectively. Although patients with different subtypes display similar symptoms, they may develop distinct diabetes-related complications and require different treatments depending on the type of the mutation. Genetic analysis of MODY patients revealed more than 400 missense/nonsense mutations in HNF1-α, HNF4-α, and HNF1-β genes, however only a small portion of them are functionally characterized. Evaluation of nonsense mutations are more direct as they lead to premature stop codons and mostly in mRNA decay or nonfunctional truncated proteins. However, interpretation of the single amino acid change (missense) mutation is not such definite, as effect of the variant may vary depending on the location and also the substituted amino acid. Mutations with benign effect on the protein function may not be the pathologic variant and further genetic testing may be required. Here, we discuss the functional characterization analysis of single amino acid change mutations identified in HNF1-α, HNF4-α, and HNF1-β genes and evaluate their roles in MODY pathogenesis. This review will contribute to comprehend HNF nuclear family-related molecular mechanisms and to develop more accurate diagnosis and treatment based on correct evaluation of pathologic effects of the variants.




Similar content being viewed by others
References
Shields BM, Hicks S, Shepherd MH et al (2010) Maturity-onset diabetes of the young (MODY): How many cases are we missing? Diabetologia 53:2504–2508. https://doi.org/10.1007/s00125-010-1799-4
Schober E, Rami B, Grabert M et al (2009) Phenotypical aspects of maturity-onset diabetes of the young (MODY diabetes) in comparison with Type 2 diabetes mellitus (T2DM) in children and adolescents: experience from a large multicentre database. Diabet Med 26:466–473. https://doi.org/10.1111/j.1464-5491.2009.02720.x
Attiya K, Sahar F (2012) Maturity-onset diabetes of the young (MODY) genes: literature review. Clinical Practice 2012:4–11. https://doi.org/10.5923/j.cp.20120101.02
Juszczak A, Owen K (2014) Identifying subtypes of monogenic diabetes Review. Diabetes Manage 4:49–61. https://doi.org/10.2217/DMT.13.59
Siddiqui K, Musambil M, Nazir N (2015) Maturity onset diabetes of the young (MODY)-History, first case reports and recent advances. Gene 555:66–71. https://doi.org/10.1016/j.gene.2014.09.062
Fajans SS, Bell GI, Polonsky KS (2001) Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med 345:971–980. https://doi.org/10.1056/nejmra002168
Hansen T, Eiberg H, Rouard M et al (1997) Novel MODY3 mutations in the hepatocyte nuclear factor-1α gene: evidence for a hyperexcitability of pancreatic β-cells to intravenous secretagogues in a glucose-tolerant carrier of a P447L mutation. Diabetes 46:726–730. https://doi.org/10.2337/diab.46.4.726
Nammo T, Yamagata K, Tanaka T et al (2008) Expression of HNF-4α (MODY1), HNF-1β (MODY5), and HNF-1α (MODY3) proteins in the developing mouse pancreas. Gene Expr Patterns 8:96–106. https://doi.org/10.1016/j.modgep.2007.09.006
Roehlen N, Hilger H, Stock F et al (2018) 17q12 Deletion Syndrome as a Rare Cause for Diabetes Mellitus Type MODY5. J Clin Endocrinol Metab 103:3601–3610. https://doi.org/10.1210/jc.2018-00955
Yamagata K, Furuta H, Oda N et al (1996) Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1). Nature 384:458–460. https://doi.org/10.1038/384458a0
Peixoto-Barbosa R, Reis AF, Giuffrida FMA (2020) Update on clinical screening of maturity-onset diabetes of the young (MODY). Diabetol Metab Syndr 12:50. https://doi.org/10.1186/s13098-020-00557-9
Delvecchio M, Pastore C, Giordano P (2020) Treatment Options for MODY Patients: A Systematic Review of Literature. Diabetes Therapy 11:1667–1685. https://doi.org/10.1007/s13300-020-00864-4
Urakami T (2019) Maturity-onset diabetes of the young (MODY): current perspectives on diagnosis and treatment. Diabetes Metab Syndrome Obes 12:1047–1056
Morita K, Saruwatari J, Tanaka T et al (2017) Common variants of HNF1A gene are associated with diabetic retinopathy and poor glycemic control in normal-weight Japanese subjects with type 2 diabetes mellitus. J Diabetes Complicat 31:483–488. https://doi.org/10.1016/j.jdiacomp.2016.06.007
Flannick J, Beer NL, Bick AG et al (2013) Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes. Nat Genet 45:1380–1387. https://doi.org/10.1038/ng.2794
Malikova J, Kaci A, Dusatkova P et al (2020) Functional analyses of hnf1a-mody variants refine the interpretation of identified sequence variants. J Clin Endocrinol Metab 105:E1377–E1386. https://doi.org/10.1210/clinem/dgaa051
Althari S, Najmi LA, Bennett AJ et al (2020) Unsupervised clustering of missense variants in HNF1A using multidimensional functional data aids clinical interpretation. Am J Hum Genet 107:670–682. https://doi.org/10.1016/j.ajhg.2020.08.016
Cereghini S (1996) Liver-enriched transcription factors and hepatocyte differentiation. FASEB J 10:267–282. https://doi.org/10.1096/fasebj.10.2.8641560
Lau HH, Ng NHJ, Loo LSW et al (2018) The molecular functions of hepatocyte nuclear factors—in and beyond the liver. J Hepatol 68:1033–1048. https://doi.org/10.1016/j.jhep.2017.11.026
Ryffel GU (2001) Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J Mol Endocrinol 27:11–29. https://doi.org/10.1677/jme.0.0270011
Galán M, García-Herrero CM, Azriel S et al (2011) Differential effects of HNF-1α mutations associated with familial young-onset diabetes on target gene regulation. Mol Med 17:256–265. https://doi.org/10.2119/molmed.2010.00097
Kim EK, Lee JS, Il CH et al (2014) Identification and functional characterization of P159L mutation in HNF1B in a family with maturity-onset diabetes of the young 5 (MODY5). Genomics Inform 12:240. https://doi.org/10.5808/gi.2014.12.4.240
Navas MA, Munoz-Elias EJ, Kim J et al (1999) Functional characterization of the MODY1 gene mutations HNF4(R127W), HNF4(V255M), and HNF4(E276Q). Diabetes 48:1459–1465. https://doi.org/10.2337/diabetes.48.7.1459
Lee SS, Cha E-Y, Jung H-J et al (2008) Genetic polymorphism of hepatocyte nuclear factor-4α influences human cytochrome P450 2D6 activity. Hepatology 48:635–645. https://doi.org/10.1002/hep.22396
Fang B, Mane-Padros D, Bolotin E et al (2012) Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors. Nucleic Acids Res 40:5343–5356. https://doi.org/10.1093/nar/gks190
Singh P, Tung SP, Han EH et al (2019) Dimerization defective MODY mutations of hepatocyte nuclear factor 4α. Mutat Res Fundam Mol Mech Mutagen 814:1–6. https://doi.org/10.1016/j.mrfmmm.2019.01.002
Thomas H, Badenberg B, Bulman M et al (2002) Evidence for haploinsufficiency of the human HNF1α gene revealed by functional characterization of MODY3-associated mutations. Biol Chem 383:1691–1700. https://doi.org/10.1515/BC.2002.190
Uhlén M, Fagerberg L, Hallström BM et al (2015) Tissue-based map of the human proteome. Science. https://doi.org/10.1126/science.1260419
Juszczak A, Gilligan LC, Hughes BA et al (2020) Altered cortisol metabolism in individuals with HNF1A-MODY. Clin Endocrinol 93:269–279. https://doi.org/10.1111/cen.14218
Scherer SE, Muzny DM, Buhay CJ et al (2006) The finished DNA sequence of human chromosome 12. Nature 440:346–351. https://doi.org/10.1038/nature04569
Anik A, Çatli G, Abaci A, Böber E (2015) Maturity-onset diabetes of the young (MODY): an update. J Pediatr Endocrinol Metab 28:251–263
Radha V, Ek J, Anuradha S et al (2009) Identification of novel variants in the hepatocyte nuclear factor-1α gene in South Indian patients with maturity onset diabetes of young. J Clin Endocrinol Metab 94:1959–1965. https://doi.org/10.1210/jc.2008-2371
Awa WL, Thon A, Raile K et al (2011) Genetic and clinical characteristics of patients with HNF1A gene variations from the German-Austrian DPV database. Eur J Endocrinol 164:513–520. https://doi.org/10.1530/EJE-10-0842
Balamurugan K, Bjørkhaug L, Mahajan S et al (2016) Structure–function studies of HNF1A (MODY3) gene mutations in South Indian patients with monogenic diabetes. Clin Genet 90:486–495. https://doi.org/10.1111/cge.12757
Bjørkhaug L, Bratland A, Njølstad PR, Molven A (2005) Functional dissection of the HNF-1alpha transcription factor: a study on nuclear localization and transcriptional activation. DNA Cell Biol 24:661–669. https://doi.org/10.1089/dna.2005.24.661
Colclough K, Bellanne-Chantelot C, Saint-Martin C et al (2013) Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha and 4 alpha in maturity-onset diabetes of the young and Hyperinsulinemic Hypoglycemia. Hum Mutat 34:669–685. https://doi.org/10.1002/humu.22279
Ikema T, Shimajiri Y, Komiya I et al (2002) Identification of three new mutations of the HNF-1α gene in Japanese MODY families. Diabetologia 45:1713–1718. https://doi.org/10.1007/s00125-002-0972-9
Sneha P, Thirumal KD, George PDC et al (2017) Determining the role of missense mutations in the POU domain of HNF1A that reduce the DNA-binding affinity: a computational approach. PLoS ONE 12:e0174953–e0174953. https://doi.org/10.1371/journal.pone.0174953
Mcguffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server
Buchan DWA, Jones DT (2019) The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res 47:W402–W407. https://doi.org/10.1093/nar/gkz297
Narayana N, Phillips NB, Hua QX et al (2006) Diabetes Mellitus due to misfolding of a β-cell transcription factor: stereospecific frustration of a Schellman Motif in HNF-1α. J Mol Biol 362:414–429. https://doi.org/10.1016/j.jmb.2006.06.086
Chi YI, Frantz JD, Oh BC et al (2002) Diabetes mutations delineate an atypical POU domain in HNF-1α. Mol Cell 10:1129–1137. https://doi.org/10.1016/S1097-2765(02)00704-9
Alvelos MI, Gonçalves CI, Coutinho E et al (2020) Maturity-onset diabetes of the young (MODY) in Portugal: novel GCK, HNFA1 and HNFA4 mutations. J Clin Med 9:288. https://doi.org/10.3390/jcm9010288
Stenson PD, Mort M, Ball EV et al (2020) The Human Gene Mutation Database (HGMD®): optimizing its use in a clinical diagnostic or research setting. Hum Genet 139:1197–1207. https://doi.org/10.1007/s00439-020-02199-3
Narayana N, Hua QX, Weiss MA (2001) The dimerization domain of HNF-1α: Structure and plasticity of an intertwined four-helix bundle with application to diabetes mellitus. J Mol Biol 310:635–658. https://doi.org/10.1006/jmbi.2001.4780
Luscombe NM, Laskowski RA, Thornton JM (2001) Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res 29:2860–2874. https://doi.org/10.1093/nar/29.13.2860
Bjørkhaug L, Sagen JV, Thorsby P et al (2003) Hepatocyte nuclear factor-1α gene mutations and diabetes in Norway. J Clin Endocrinol Metab 88:920–931. https://doi.org/10.1210/jc.2002-020945
Ban N, Yamada Y, Someya Y et al (2002) Hepatocyte nuclear factor-1α recruits the transcriptional co-activator p300 on the GLUT2 gene promoter. Diabetes 51:1409–1418. https://doi.org/10.2337/diabetes.51.5.1409
Fareed FMA, Korulu S, Özbil M, Çapan ÖY (2021) HNF1A-MODY mutations in nuclear localization signal impair HNF1A-import receptor KPNA6 interactions. Protein J. https://doi.org/10.1007/s10930-020-09959-0
Triggs-Raine BL, Kirkpatrick RD, Kelly SL et al (2002) HNF-1α G319S, a transactivation-deficient mutant, is associated with altered dynamics of diabetes onset in an Oji-Cree community. Proc Natl Acad Sci USA 99:4614–4619. https://doi.org/10.1073/pnas.062059799
Vaxillaire M, Rouard M, Yamagata K et al (1997) Identification of nine novel mutations in the hepatocyte nuclear factor 1 alpha gene associated with maturity-onset diabetes of the young (MODY3). Hum Mol Genet 6:583–586. https://doi.org/10.1093/hmg/6.4.583
Yamada S, Tomura H, Nishigori H et al (1999) Identification of mutations in the hepatocyte nuclear factor-1α gene in Japanese subjects with early-onset NIDDM and functional analysis of the mutant proteins. Diabetes 48:645–648. https://doi.org/10.2337/diabetes.48.3.645
Xu JY, Chan V, Zhang WY et al (2002) Mutations in the hepatocyte nuclear factor-1α gene in Chinese MODY families: prevalence and functional analysis. Diabetologia 45:744–746. https://doi.org/10.1007/s00125-002-0814-9
Holmkvist J, Cervin C, Lyssenko V et al (2006) Common variants in HNF-1 α and risk of type 2 diabetes. Diabetologia 49:2882–2891. https://doi.org/10.1007/s00125-006-0450-x
Cervin C, Orho-Melander M, Ridderstråle M et al (2002) Characterisation of a naturally occurring mutation “L107I” in the HNF1α “MODY3” gene. Diabetologia 45:1703–1708. https://doi.org/10.1007/s00125-002-0977-4
Najmi LA, Aukrust I, Flannick J et al (2017) Functional investigations of HNF1A identify rare variants as risk factors for type 2 diabetes in the general population. Diabetes 66:335–346. https://doi.org/10.2337/db16-0460
Vaxillaire M, Abderrahmani A, Boutin P et al (1999) Anatomy of a homeoprotein revealed by the analysis of human MODY3 mutations. J Biol Chem 274:35639–35646. https://doi.org/10.1074/jbc.274.50.35639
Rozenkova K, Malikova J, Nessa A et al (2015) High incidence of heterozygous ABCC8 and HNF1A mutations in Czech patients with congenital hyperinsulinism. J Clin Endocrinol Metab 100:E1540–E1549. https://doi.org/10.1210/jc.2015-2763
Yoshiuchi I, Yamagata K, Yang Q et al (1999) Three new mutations in the hepatocyte nuclear factor-1α gene in Japanese subjects with diabetes mellitus: clinical features and functional characterization. Diabetologia 42:621–626. https://doi.org/10.1007/s001250051204
Plengvidhya N, Boonyasrisawat W, Chongjaroen N et al (2009) Mutations of maturity-onset diabetes of the young (MODY) genes in Thais with early-onset type 2 diabetes mellitus: ORIGINAL ARTICLE. Clin Endocrinol 70:847–853. https://doi.org/10.1111/j.1365-2265.2008.03397.x
Bjorkhaug L, Ye H, Horikawa Y et al (2000) MODY associated with two novel hepatocyte nuclear factor-1α loss-of-function mutations (P112L and Q466X). Biochem Biophys Res Commun 279:792–798. https://doi.org/10.1006/bbrc.2000.4024
Deloukas P, Matthews LH, Ashurst J et al (2001) The DNA sequence and comparative analysis of human chromosome 20. Nature 414:865–871. https://doi.org/10.1038/414865a
Huang J, Levitsky LL, Rhoads DB (2009) Novel P2 promoter-derived HNF4α isoforms with different N-terminus generated by alternate exon insertion. Exp Cell Res 315:1200–1211. https://doi.org/10.1016/j.yexcr.2009.01.004
Ko HL, Zhuo Z, Chee E, Correspondence R (2019) HNF4α; combinatorial isoform heterodimers activate distinct gene targets that differ from their corresponding homodimers. Cell Rep 26:2549-2557.e3. https://doi.org/10.1016/j.celrep.2019.02.033
Tanaka T, Jiang S, Hotta H et al (2006) Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4α in the pathogenesis of human cancer. J Pathol 208:662–672. https://doi.org/10.1002/path.1928
Fukushima-Uesaka H, Saito Y, Maekawa K et al (2006) Novel genetic variations and haplotypes of hepatocyte nuclear factor 4alpha (HNF4A) found in Japanese type II diabetic patients. Drug Metab Pharmacokinet 21:337–346. https://doi.org/10.2133/dmpk.21.337
Evans RM, Mangelsdorf DJ (2014) Nuclear receptors, RXR, and the big bang. Cell 157:255–266. https://doi.org/10.1016/j.cell.2014.03.012
Marchesin V, Pérez-Martí A, Le Meur G et al (2019) Molecular basis for autosomal-dominant Renal Fanconi Syndrome caused by HNF4A. Cell Rep 29:4407-4421.e5. https://doi.org/10.1016/j.celrep.2019.11.066
Sladek FM, Zhong W, Lai E, Darnell JE (1990) Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev 4:2353–2365. https://doi.org/10.1101/gad.4.12b.2353
Costa RH, Kalinichenko VV, Holterman AXL, Wang X (2003) Transcription factors in liver development, differentiation, and regeneration. Hepatology 38:1331–1347. https://doi.org/10.1016/j.hep.2003.09.034
Dhe-Paganon S, Duda K, Iwamoto M et al (2002) Crystal structure of the HNF4α ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem 277:37973–37976. https://doi.org/10.1074/jbc.C200420200
Chandra V, Huang P, Potluri N et al (2013) Multidomain integration in the structure of the HNF-4α nuclear receptor complex. Nature 495:394–398. https://doi.org/10.1038/nature11966
Lu P, Rha GB, Melikishvili M et al (2008) Structural basis of natural promoter recognition by a unique nuclear receptor, HNF4α: diabetes gene product. J Biol Chem 283:33685–33697. https://doi.org/10.1074/jbc.M806213200
Han EH, Singh P, Lee IK et al (2019) ErbB3-binding protein 1 (EBP1) represses HNF4α-mediated transcription and insulin secretion in pancreatic β-cells. J Biol Chem 294:13983–13994. https://doi.org/10.1074/jbc.RA119.009558
Clemente M, Vargas A, Ariceta G et al (2017) Hyperinsulinaemic hypoglycaemia, renal Fanconi syndrome and liver disease due to a mutation in the HNF4A gene. Endocrinol Diabetes Metab Case Rep. https://doi.org/10.1530/edm-16-0133
Liu J, Shen Q, Li G, Xu H (2018) HNF4A-related Fanconi syndrome in a Chinese patient: a case report and review of the literature. J Med Case Rep 12:203. https://doi.org/10.1186/s13256-018-1740-x
Oxombre B, Kouach M, Moerman E et al (2004) The G115S mutation associated with maturity-onset diabetes of the young impairs hepatocyte nuclear factor 4α activities and introduces a PKA phosphorylation site in its DNA-binding domain. Biochem J 383:573–580. https://doi.org/10.1042/BJ20040473
Sun Z, Xu Y (2020) Nuclear receptor coactivators (NCOAs) and corepressors (NCORs) in the brain. Endocrinology 161:1–12. https://doi.org/10.1210/endocr/bqaa083
Eeckhoute J, Formstecher P, Laine B (2004) Hepatocyte nuclear factor 4α enhances the hepatocyte nuclear factor 1α-mediated activation of transcription. Nucleic Acids Res 32:2586–2593. https://doi.org/10.1093/nar/gkh581
Pearson ER, Pruhova S, Tack CJ et al (2005) Molecular genetics and phenotypic characteristics of MODY caused by hepatocyte nuclear factor 4α mutations in a large European collection. Diabetologia 48:878–885. https://doi.org/10.1007/s00125-005-1738-y
Taniguchi H, Fujimoto A, Kono H et al (2018) Loss-of-function mutations in Zn-finger DNA-binding domain of HNF4A cause aberrant transcriptional regulation in liver cancer. Oncotarget 9:26144–26156
Oxombre B, Moerman E, Eeckhoute J et al (2002) Mutations in hepatocyte nuclear factor 4α (HNF4α) gene associated with diabetes result in greater loss of HNF4α function in pancreatic β-cells than in nonpancreatic β-cells and in reduced activation of the apolipoprotein CIII promoter in hepatic cells. J Mol Med 80:423–430. https://doi.org/10.1007/s00109-002-0340-8
Yang Y, Zhou T-C, Liu Y-Y et al (2016) Identification of HNF4A Mutation p. T130I and HNF1A Mutations p.I27L and p.S487N in a Han Chinese Family with Early-Onset Maternally Inherited Type 2 Diabetes. J Diabetes Res 2016:1–8. https://doi.org/10.1155/2016/3582616
Ek J, Rose CS, Jensen DP et al (2005) The functional Thr130Ile and Val255Met polymorphisms of the hepatocyte nuclear factor-4α (HNF4A): gene associations with type 2 diabetes or altered β-cell function among danes. J Clin Endocrinol Metab 90:3054–3059. https://doi.org/10.1210/jc.2004-2159
Adalat S, Woolf AS, Johnstone KA et al (2009) HNF1B mutations associate with hypomagnesemia and renal magnesium wasting. J Am Soc Nephrol 20:1123–1131. https://doi.org/10.1681/ASN.2008060633
Bártů M, Hojný J, Hájková N et al (2020) Analysis of expression, epigenetic, and genetic changes of HNF1B in 130 kidney tumours. Sci Rep 10:1–11. https://doi.org/10.1038/s41598-020-74059-z
Zody MC, Garber M, Adams DJ et al (2006) DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature 440:1045–1049. https://doi.org/10.1038/nature04689
Chan SC, Zhang Y, Shao A et al (2018) Mechanism of fibrosis in HNF1B-related autosomal dominant tubulointerstitial kidney disease. J Am Soc Nephrol 29:2493–2509. https://doi.org/10.1681/ASN.2018040437
Ferrè S, Igarashi P (2019) New insights into the role of HNF-1β in kidney (patho)physiology. Pediatr Nephrol 34:1325–1335. https://doi.org/10.1007/s00467-018-3990-7
Nakayama M, Nozu K, Goto Y et al (2010) HNF1B alterations associated with congenital anomalies of the kidney and urinary tract. Pediatr Nephrol 25:1073–1079. https://doi.org/10.1007/s00467-010-1454-9
Painter JN, O’Mara TA, Batra J et al (2015) Fine-mapping of the HNF1B multicancer locus identifies candidate variants that mediate endometrial cancer risk. Hum Mol Genet 24:1478–1492. https://doi.org/10.1093/hmg/ddu552
Clissold RL, Hamilton AJ, Hattersley AT et al (2015) HNF1B-associated renal and extra-renal disease—an expanding clinical spectrum. Nat Rev Nephrol 11:102–112. https://doi.org/10.1038/nrneph.2014.232
Heidet L, Decramer S, Pawtowski A et al (2010) Spectrum of HNF1B mutations in a large cohort of patients who harbor renal diseases. Clin J Am Soc Nephrol 5:1079–1090. https://doi.org/10.2215/CJN.06810909
Hojny J, Bartu M, Krkavcova E et al (2020) Identification of novel HNF1B mRNA splicing variants and their qualitative and semi-quantitative profile in selected healthy and tumour tissues. Sci Rep 10:1–11. https://doi.org/10.1038/s41598-020-63733-x
Lu P, Geun BR, Chi YI (2007) Structural basis of disease-causing mutations in hepatocyte nuclear factor 1β. Biochemistry 46:12071–12080. https://doi.org/10.1021/bi7010527
Barbacci E, Chalkiadaki A, Masdeu C et al (2004) HNF1β/TCF2 mutations impair transactivation potential through altered co-regulator recruitment. Hum Mol Genet 13:3139–3149. https://doi.org/10.1093/hmg/ddh338
Edghill EL, Bingham C, Ellard S, Hattersley AT (2006) Mutations in hepatocyte nuclear factor-1β and their related phenotypes. J Med Genet 43:84–90. https://doi.org/10.1136/jmg.2005.032854
Kitanaka S, Miki Y, Hayashi Y, Igarashi T (2004) Promoter-Specific Repression of hepatocyte nuclear factor (HNF)-1β and HNF-1α transcriptional activity by an HNF-1β missense mutant associated with type 5 maturity-onset diabetes of the young with hepatic and biliary manifestations. J Clin Endocrinol Metab 89:1369–1378. https://doi.org/10.1210/jc.2003-031308
Okonechnikov K, Golosova O, Fursov M (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. https://doi.org/10.1093/bioinformatics/bts091
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Çubuk, H., Yalçın Çapan, Ö. A Review of Functional Characterization of Single Amino Acid Change Mutations in HNF Transcription Factors in MODY Pathogenesis. Protein J 40, 348–360 (2021). https://doi.org/10.1007/s10930-021-09991-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-021-09991-8