Abstract
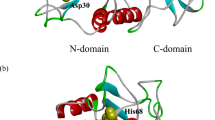
The crystal structure of the S189D+A226G rat chymotrypsin-B mutant has been determined at 2.2 Å resolution. This mutant is the most trypsin-like mutant so far in the line of chymotrypsin-to-trypsin conversions, aiming for a more complete understanding of the structural basis of substrate specificity in pancreatic serine proteases. A226G caused significant rearrangements relative to S189D chymotrypsin, allowing an internal conformation of Asp189 which is close to that in trypsin. Serious distortions remain, however, in the activation domain, including zymogen-like features. The pH-profile of activity suggests that the conformation of the S1–site of the mutant is influenced also by the P1 residue of the substrate.




Similar content being viewed by others
Abbreviations
- AMC:
-
Amino-4-Methyl Coumarin
- BPTI:
-
Bovine pancreatic trypsin inhibitor
- CABS:
-
4-(Cyclohexylamino)-1-butanesulfonic acid
- CCP4:
-
Collaborative Computational Project Number 4
- CHES:
-
2-(Cyclohexylamino)ethanesulfonic acid
- DPI:
-
Dispersion precision indicator
- ESRF:
-
European Synchrotron Radiation Facility
- HEPES:
-
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid
- MES:
-
2-(N-Morpholino)ethanesulfonic acid
- MOPS:
-
3-(N-Morpholino)propanesulfonic acid
- MUGB:
-
4-Guanidinobenzoic acid 4-methylumbelliferyl ester hydrochloride
- PDB:
-
Protein data bank
- PEG:
-
Polyethylene glycol
- SBTI:
-
Soybean trypsin inhibitor
- SDS-PAGE:
-
Sodium dodecyl sulfate - polyacrylamide gel electrophoresis
- TLS:
-
Translational, librational, screw
- TRIS:
-
Tris(hydroxymethyl)aminomethane
References
Schechter I, Berger A (1967) Biochem Biophys Res Commun 27:157–162
Gráf L, Jancsó A, Szilágyi L, Hegyi Gy, Pintér K, Náray-Szabó G, Hepp J, Medzihradszky K, Rutter WJ (1988) Proc Natl Acad Sci USA 85:4961–4965
Hedstrom L, Szilágyi L, Rutter WJ (1992) Science 255:1249–1253
Hedstrom L, Perona JJ, Rutter WJ (1994) Biochemistry 33:8757–8763
Hedstrom L, Farr Jones S, Kettner CA, Rutter WJ (1994) Biochemistry 33:8764–8769
Gráf L (1995) In: Zwilling R (ed) Natural sciences and human thought. Springer-Verlag, Berlin, Heidelberg, pp 139–148
Steitz TA, Henderson R, Blow DM (1969) J Mol Biol 46:337–348
Venekei I, Szilágyi L, Gráf L, Rutter WJ (1996) FEBS Lett 379:143–147
Craik CS, Largman C, Fletcher T, Roczniak S, Barr PJ, Fletterick RJ, Rutter WJ (1985) Science 228:291–297
Wilke ME, Higaki JN, Craik CS, Fletterick RJ (1991) J Mol Biol 219:525–532
Jelinek B, Antal J, Venekei I, Gráf L (2004) Protein Eng Des Sel 17:127–131
Jameson GW, Adams DV, Kyle WS, Elmore DT (1973) Biochem J 131:107–117
CCP 4 (1994) Acta Cryst D 50:760–763
Navaza J (2001) Acta Crystallogr D Biol Crystallogr 57:1367–1372
Szabó E, Venekei I, Böcskei Zs, Náray-Szabó G, Gráf L (2003) J Mol Biol 331(5):1121–1130
Terwilliger TC (2000) Acta Cryst D56:965–972
Emsley P, Cowtan K (2004) Acta Cryst D60:2126–2132
Murshudov GN, Vagin AA, Dodson EJ (1997) Acta Cryst D53:240–255
Hooft RW, Vriend G, Sander C, Abola EE (1996) Nature 381:272
Laskowski RA, MAcArtur MW, Moss DS, Thornton JM (1993) J Appl Crystallog 26:283–291
Luzzati PV (1952) Acta Cryst 5:802–810
Cruickshank DWJ (1999) Acta Cryst D55:583–601
Lesk AM (1991) Protein architecture: a practical guide. IRL Press, Oxford
Madsen D, Kleywegt GJ (2002) J Appl Crystallog 35:137–139
Marquart M, Walter J, Deisenhofer J, Bode W, Huber R (1983) Acta Crystallogr Sect B v39:480
Brady K, Wei AZ, Ringe D, Abeles RH (1990) Biochemistry v29:7600–7607
Segel IH (1993) Enzyme Kinetics: Behavior and analysis of rapid equilibrium and steady-state enzyme systems, Wiley Classics Library Edition
Dementiev A, Dobó J, Gettins PGW (2006) J Biol Chem 281(6):3452–3457
Katz B, Kossiakoff A (1986) J Biol Chem 261(33):15480-15485
Fodor K, Harmat V, Neutze R, Szilágyi L, Gráf L, Katona G (2006) Biochemistry 21;45(7):2114–2121
Radisky ES, Lee JM, Lu CJ, Koshland DE Jr (2006) Proc Natl Acad Sci U S A 103(18):6835–6840
Fuhrmann CN, Daugherty MD, Agard DA (2006) J Am Chem Soc 128(28):9086–9102
Liu B, Schofield CJ, Wilmouth RC (2006) J Biol Chem 281(33):24024–24035
Fersht AR (1972) J Mol Biol 64(2):497–509
Verheyden G, Matrai J, Volckaert G, Engelborghs Y (2004) Prot Sci 13:2533–2540
Szabó E, Böcskei Zs, Náray-Szabó G, Gráf L (1999) Eur J Biochem 263(1):20–26
Bode W, Schwager P, Huber R (1978) J Mol Biol 118:99–112
Acknowledgements
We are grateful for the beamline staff at the ESRF ID14–2 beamline for their expert assistance. This work was supported by the Hungarian Research Fund OTKA TS049812 and T047154 to L.G., G. K. acknowledges the support from EMBO and CEA.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jelinek, B., Katona, G., Fodor, K. et al. The Crystal Structure of a Trypsin-like Mutant Chymotrypsin: The Role of Position 226 in the Activity and Specificity of S189D Chymotrypsin. Protein J 27, 79–87 (2008). https://doi.org/10.1007/s10930-007-9110-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-007-9110-3