Abstract
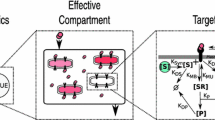
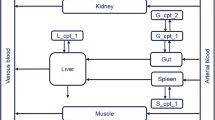
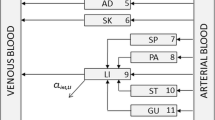
We present a Bayesian automated method to reduce by lumping, a large system described by differential equations which takes into account parameter variability. Model reduction is a potentially useful tool to simplify large systems but suffers from lack of robustness over the model parameter values. With the present method we address this problem by incorporating a prior parameter distribution in the determination of the optimal lumping scheme in a Bayesian manner. Applications of this method may include PBPK models for the drug distribution and/or Systems Biology models for the drug action. The method builds on our previously published algorithm for lumping that works stepwise, reducing the system’s dimension by one at each step and where each successive step is conditional to the previous ones. We applied the methodology to a PBPK model for barbiturates taken from the literature. An arbitrary variability of 20% CV was added to the nominal reported parameter values. The Bayesian method performed better than the method which ignored the parameter variability, producing a lumping scheme which, while not optimal for any parameter value, was optimal on average. On the other hand the simple, non-Bayesian method produced a lumping scheme which while optimal for the nominal parameter values, was very poor for most other values within the prior distribution. Further, we discuss the generality of a lumping strategy to reduce a model and we argue that this is more powerful than elimination of states, with the latter being almost a special case of lumping.





Similar content being viewed by others
References
Blakey GE, Nestorov IA, Arundel PA, Aarons LJ, Rowland M (1997) Quantitative structure-pharmacokinetics relationships: I. Development of a whole-body physiologically based model to characterize changes in pharmacokinetics across a homologous series of barbiturates in the rat. J Pharmacokinet Biopharm 25:277–312
Yangm K, Bai H, Ouyang Q, Lai L, Tang C (2008) Finding multiple target optimal intervention in disease-related molecular network. Mol Syst Biol 4:228
Okino MS, Mavrovouniotis ML (1998) Simplification of mathematical models of chemical reaction systems. Chem Rev 98:391–408
Wei J, Kuo JCW (1969) A lumping analysis in monomolecular reaction systems—analysis of exactly lumpable system. Ind Eng Chem Fund 8:114–123
Astarita G, Ocone R (1988) Lumping nonlinear kinetics. AICHE J 34:1299–1309
Li G, Rabitz H (1989) A general-analysis of exact lumping in chemical-kinetics. Chem Eng Sci 44:1413–1430
Li G, Rabitz H (1990) A general-analysis of approximate lumping in chemical-kinetics. Chem Eng Sci 45:977–1002
Li GY, Rabitz H (1991) A general lumping analysis of a reaction system coupled with diffusion. Chem Eng Sci 46:2041–2053
Nestorov IA, Aarons LJ, Arundel PA, Rowland M (1998) Lumping of whole-body physiologically based pharmacokinetic models. J Pharmacokinet Biopharm 26:21–46
Brochot C, Toth J, Bois FY (2005) Lumping in pharmacokinetics. J Pharmacokinet Pharmacodyn 32:719–736
Dokoumetzidis A, Aarons L (2009) Proper lumping in systems biology models. IET Syst Biol 3:40–51
Gillespie DT, Cao Y, Sanft KR, Petzold LR (2009) The subtle business of model reduction for stochastic chemical kinetics. J Chem Phys 130:064103
Radulescu O, Gorban AN, Zinovyev A, Lilienbaum A (2008) Robust simplifications of multiscale biochemical networks. BMC Syst Biol 2:86
Gueorguieva I, Nestorov IA, Rowland M (2006) Reducing whole body physiologically based pharmacokinetic models using global sensitivity analysis: diazepam case study. J Pharmacokinet Pharmacodyn 33:1–27
McKay MD, Conover WJ, Beckman RJ (1979) A comparison of three methods for selecting values of input variables in the analysis of output from a computer code. Technometrics 21:239–245
Acknowledgment
This work was conducted while AD was employed by the School of Pharmacy of the University of Manchester and was funded by a grant from Novartis.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dokoumetzidis, A., Aarons, L. A method for robust model order reduction in pharmacokinetics. J Pharmacokinet Pharmacodyn 36, 613–628 (2009). https://doi.org/10.1007/s10928-009-9141-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-009-9141-9