Abstract
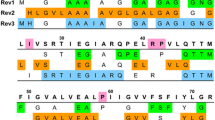
Solid-state Nuclear Magnetic Resonance (ssNMR) is an emerging technique to investigate the structures and dynamics of membrane proteins in an artificial or native membrane environment. However, the structural studies of proteins by ssNMR are usually prolonged or impeded by signal assignments, especially the assignments of signals for collection of distance restraints, because of serious overlapping of signals in 2D 13C–13C spectra. Sparse labeling of 13C spins is an effective approach to simplify the 13C spectra and facilitate the extractions of distance restraints. Here, we propose a new reverse labeling combination of six types of amino acid residues (Ile, Leu, Phe, Trp, Tyr and Lys), and show a clean reverse labeling effect on a model membrane protein E. coli aquaporin Z (AqpZ). We further combine this reverse labeling combination and alternate 13C–12C labeling, and demonstrate an enhanced dilution effect in 13C–13C spectra. In addition, the influences of reverse labeling on the labeling of the other types of residues are quantitatively analyzed in the two strategies (1, reverse labeling and 2, reverse labeling combining alternate 13C–12C labeling). The signal intensities of some other types of residues in 2D 13C–13C spectra are observed to be 20–50% weaker because of the unwanted reverse labeling. The extensively sparse 13C labeling proposed in this study is expected to be useful in the collection of distance restraints using 2D 13C–13C spectra of membrane proteins.






Similar content being viewed by others
References
Atreya HS (2012) Isotope labeling in biomolecular NMR preface. Adv Exp Med Biol 992:V
Banigan JR et al (2013) Combination of 15 N reverse labeling and afterglow spectroscopy for assigning membrane protein spectra by magic-angle-spinning solid-state NMR: application to the multidrug resistance protein. EmrE J Biomol NMR 55:391–399
Bayro MJ et al (2010) High-resolution MAS NMR analysis of PI3-SH3 amyloid fibrils: backbone conformation and implications for protofilament assembly and structure. Biochemistry 49:7474–7484
Berger C et al (2013) Efficient isotopic tryptophan labeling of membrane proteins by an indole controlled process conduct. Biotechnol Bioeng 110:1681–1690
Castellani F et al (2002) Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature 420:98–102
Crowhurst KA et al (2003) Aromatic and methyl NOES highlight hydrophobic clustering in the unfolded state of an SH3 domain. Biochemistry 42:8687–8695
Delaglio F et al (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Ding X et al (2020) De novo resonance assignment of the transmembrane domain of LR11/SorLA in E. Fcoli membranes. J Magn Reson 310:106639
Doležal M et al (2015) Resonance assignments of the myristoylated Y28F/Y67F mutant of the Mason-Pfizer monkey virus matrix protein. Biomol NMR Assign 9:229–233
Eddy MT et al (2013) Selectively dispersed isotope labeling for protein structure determination by magic angle spinning NMR. J Biomol NMR 57:129–139
Etzkorn M et al (2007) Secondary structure, dynamics, and topology of a seven-helix receptor in native membranes, studied by solid-state NMR spectroscopy. Angew Chem Int Ed Engl 46:459–462
Fan Y et al (2015) Isotope labeling of eukaryotic membrane proteins in yeast for solid-state NMR. Methods Enzymol 565:193–212
Fasshuber HK et al (2015) Specific C-13 labeling of leucine, valine and isoleucine methyl groups for unambiguous detection of long-range restraints in protein solid-state NMR studies. J Magn Reson 252:10–19
Heise H et al (2005) Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA 102:15871–15876
Higman VA et al (2009) Assigning large proteins in the solid state: a MAS NMR resonance assignment strategy using selectively and extensively C-13-labelled proteins. J Biomol NMR 44:245–260
Hiller M et al (2008) [2,3-C-13]-labeling of aromatic residues-getting a head start in the magic-angle-spinning NMR assignment of membrane proteins. J Am Chem Soc 130:408
Hong M (1999) Determination of multiple phi-torsion angles in proteins by selective and extensive C-13 labeling and two-dimensional solid-state NMR. J Magn Reson 139:389–401
Hong M et al (1999) Selective and extensive 13C labeling of a membrane protein for solid-state NMR investigations. J Biomol NMR 14:71–74
Kang S-J et al (2018) Direct assignment of 13 C solid-state NMR signals of TF o F 1 ATP synthase subunit c-ring in lipid membranes and its implication for the ring structure. J Biomol NMR 70:53–65
Goddard TD et al SPARKY 3 University of California, San Francisco
Krishnarjuna B et al (2011) Amino acid selective unlabeling for sequence specific resonance assignments in proteins. J Biomol NMR 49:39–51
Kuboniwa H et al (1995) Solution structure of calcium-free calmodulin. Nat Struct Biol 2:768–776
Kunert B et al (2014) Efficient and stable reconstitution of the ABC transporter BmrA for solid-state NMR studies. Front Mol Biosci 1:5
Lacabanne D et al (2018) Selective labeling and unlabeling strategies in protein solid-state NMR spectroscopy. J Biomol NMR 71:141–150
Lacabanne D et al (2019) Flexible-to-rigid transition is central for substrate transport in the ABC transporter BmrA from Bacillus Subtilis. Commun Biol 2:149
LeMaster DM et al (1996) Dynamical mapping of E-coli thioredoxin via C-13 NMR relaxation analysis. J Am Chem Soc 118:9255–9264
Lin MT et al (2011) A rapid and robust method for selective isotope labeling of proteins. Methods 55:370–378
Lin MT et al (2015) Escherichia coli auxotroph host strains for amino acid-selective isotope labeling of recombinant proteins. Methods Enzymol 565:45–66
Loquet A et al (2010) Supramolecular interactions probed by C-13-C-13 solid-state NMR spectroscopy. J Am Chem Soc 132:15164–15166
Loquet A et al (2011) C-13 spin dilution for simplified and complete solid-state NMR resonance assignment of insoluble biological assemblies. J Am Chem Soc 133:4722–4725
Loquet A et al (2012) Atomic model of the type III secretion system needle. Nature 486:276
Lundstrom P et al (2007) Fractional C-13 enrichment of isolated carbons using [1-C-13]- or [2-C-13]-glucose facilitates the accurate measurement of dynamics at backbone C-alpha and side-chain methyl positions in proteins. J Biomol NMR 38:199–212
Marley J et al (2001) A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR 20:71–75
Morcombe CR et al (2003) Chemical shift referencing in MAS solid state NMR. J Magn Reson 162:479–486
Prasanna C et al (2015) Amino acid selective unlabeling in protein NMR spectroscopy. Method Enzymol 565:167–189
Retel JS et al (2017) Structure of outer membrane protein G in lipid bilayers. Nat Commun 8:2073
Schneider R et al (2008) Solid-state NMR spectroscopy applied to a chimeric potassium channel in lipid bilayers. J Am Chem Soc 130:7427–7435
Shi L et al (2009a) Solid-state NMR study of proteorhodopsin in the lipid environment: secondary structure and dynamics. Biochem Biophys Acta 1788:2563–2574
Shi L et al (2009b) Three-dimensional solid-state NMR study of a seven-helical integral membrane proton pump-structural insights. J Mol Biol 386:1078–1093
Sperling LJ et al (2010) Assignment strategies for large proteins by magic-angle spinning NMR: the 21-kDa disulfide-bond-forming enzyme DsbA. J Mol Biol 399:268–282
Takegoshi K et al (2001) 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett 344:631–637
Wang S et al (2013) Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nat Methods 10:1007–1012
Wang SL et al (2014) Recent advances in magic angle spinning solid state NMR of membrane proteins. Prog Nucl Mag Res Spectrosc 82:1–26
Wang SL et al (2018) Application of paramagnetic relaxation enhancements to accelerate the acquisition of 2D and 3D solid-state NMR spectra of oriented membrane proteins. Methods 138:54–61
Weininger U et al (2014) Off-resonance rotating-frame relaxation dispersion experiment for C-13 in aromatic side chains using L-optimized TROSY-selection. J Biomol NMR 59:23–29
Weininger U et al (2012) (1)(3)C relaxation experiments for aromatic side chains employing longitudinal- and transverse-relaxation optimized NMR spectroscopy. J Biomol NMR 53:181–190
Wiegand T et al (2017) Solid-state NMR and EPR spectroscopy of Mn(2+) -substituted ATP-fueled protein engines. Angew Chem Int Ed Engl 56:3369–3373
Xie H et al (2018) Solid-state NMR chemical shift assignments of aquaporin Z in lipid bilayers. Biomol NMR Assign 12:323–328
Zhao Y et al (2018) Gating mechanism of aquaporin Z in synthetic bilayers and native membranes revealed by solid-state NMR spectroscopy. J Am Chem Soc 140:7885–7895
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (Grant Nos. 2016YFA0501200 and 2017YFA0505400) and the National Natural Science Foundation of China (Grant Nos. 31600625, 21425523, 31770798, 21927801, 21991080, 21921004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tong, Q., Tan, H., Li, J. et al. Extensively sparse 13C labeling to simplify solid-state NMR 13C spectra of membrane proteins. J Biomol NMR 75, 245–254 (2021). https://doi.org/10.1007/s10858-021-00372-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-021-00372-y